2LVK
 
 | | Solution structure of Ca-bound Phl p 7 | | Descriptor: | CALCIUM ION, Polcalcin Phl p 7 | | Authors: | Henzl, M.T, Sirianni, A.G, Tanner, J.J. | | Deposit date: | 2012-07-05 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of polcalcin Phl p 7 in three ligation states: Apo-, hemi-Mg(2+) -bound, and fully Ca(2+) -bound.
Proteins, 81, 2013
|
|
2HJL
 
 | | Crystal Structure of HLA-B5703 and HIV-1 peptide | | Descriptor: | Beta-2-microglobulin, Gag protein, HLA class I histocompatibility antigen B-57 | | Authors: | Gillespie, G.M.A, Stewart-Jones, G, Rengasamy, J, Beattie, T, Bwayo, J.J, Plummer, F.A. | | Deposit date: | 2006-06-30 | | Release date: | 2006-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Strong TCR Conservation and Altered T Cell Cross-Reactivity Characterize a B*57-Restricted Immune Response in HIV-1 Infection.
J.Immunol., 177, 2006
|
|
2HHT
 
 | | C:O6-methyl-guanine pair in the polymerase-2 basepair position | | Descriptor: | 5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*CP*TP*CP*G)-3', 5'-D(*GP*TP*AP*CP*(6OG)P*AP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3', DNA polymerase I, ... | | Authors: | Warren, J.J, Forsberg, L.J, Beese, L.S. | | Deposit date: | 2006-06-28 | | Release date: | 2006-12-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structural basis for the mutagenicity of O6-methyl-guanine lesions.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FO1
 
 | | Crystal Structure of the CSL-Notch-Mastermind ternary complex bound to DNA | | Descriptor: | 5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3', 5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3', Lin-12 and glp-1 phenotype protein 1, ... | | Authors: | Wilson, J.J, Kovall, R.A. | | Deposit date: | 2006-01-12 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA.
Cell(Cambridge,Mass.), 124, 2006
|
|
5KOW
 
 | | Structure of rifampicin monooxygenase | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Pentachlorophenol 4-monooxygenase | | Authors: | Tanner, J.J, Liu, L.-K. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the Antibiotic Deactivating, N-hydroxylating Rifampicin Monooxygenase.
J.Biol.Chem., 291, 2016
|
|
2FV4
 
 | |
8PHY
 
 | |
2MUS
 
 | | HADDOCK calculated model of LIN5001 bound to the HET-s amyloid | | Descriptor: | 3''',4'-bis(carboxymethyl)-2,2':5',2'':5'',2''':5''',2''''-quinquethiophene-5,5''''-dicarboxylic acid, Heterokaryon incompatibility protein s | | Authors: | Hermann, U.S, Schuetz, A.K, Shirani, H, Saban, D, Nuvolone, M, Huang, D.H, Li, B, Ballmer, B, Aslund, A.K.O, Mason, J.J, Rushing, E, Budka, H, Hammarstrom, P, Bockmann, A, Caflisch, A, Meier, B.H, Nilsson, P.K.R, Hornemann, S, Aguzzi, A. | | Deposit date: | 2014-09-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-based drug design identifies polythiophenes as antiprion compounds.
Sci Transl Med, 7, 2015
|
|
6I93
 
 | |
5L06
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 3rd Position | | Descriptor: | DNA (5'-D(*CP*GP*(5CM)P*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Miears, H.L, Hoppins, J.J, Gruber, D.R, Kasymov, R.D, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-07-26 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
2FR4
 
 | | Structure of Fab DNA-1 complexed with a stem-loop DNA ligand | | Descriptor: | 5'-D(*CP*TP*GP*CP*CP*TP*TP*CP*AP*G)-3', TETRAETHYLENE GLYCOL, antibody heavy chain FAB, ... | | Authors: | Tanner, J.J, Ou, Z. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Impact of DNA hairpin folding energetics on antibody-ssDNA association.
J.Mol.Biol., 374, 2007
|
|
6I95
 
 | |
6I94
 
 | |
5KJY
 
 | | Co-crystal structure of PKA RI alpha CNB-B mutant (G316R/A336T) with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Lorenz, R, Moon, E, Kim, J.J, Huang, G.Y, Kim, C, Herberg, F.W. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of PKA cyclic nucleotide-binding domains reveal novel aspects of cyclic nucleotide selectivity.
Biochem. J., 474, 2017
|
|
8PHE
 
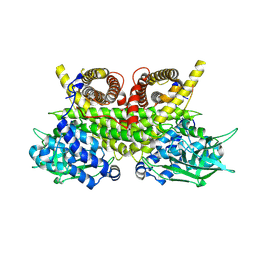 | | ACAD9-WT in complex with ECSIT-CTER | | Descriptor: | Complex I assembly factor ACAD9, mitochondrial, Evolutionarily conserved signaling intermediate in Toll pathway | | Authors: | McGregor, L, Acajjaoui, S, Desfosses, A, Saidi, M, Bacia-Verloop, M, Schwarz, J.J, Juyoux, P, Von Velsen, J, Bowler, M.W, McCarthy, A, Kandiah, E, Gutsche, I, Soler-Lopez, M. | | Deposit date: | 2023-06-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination.
Nat Commun, 14, 2023
|
|
8PHF
 
 | | Cryo-EM structure of human ACAD9-S191A | | Descriptor: | Complex I assembly factor ACAD9, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McGregor, L, Acajjaoui, S, Desfosses, A, Saidi, M, Bacia-Verloop, M, Schwarz, J.J, Juyoux, P, Von Velsen, J, Bowler, M.W, McCarthy, A, Kandiah, E, Gutsche, I, Soler-Lopez, M. | | Deposit date: | 2023-06-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination.
Nat Commun, 14, 2023
|
|
8PD0
 
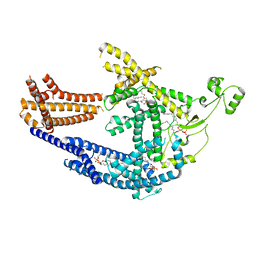 | | cryo-EM structure of Doa10 in MSP1E3D1 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | Authors: | Botsch, J.J, Braeuning, B, Schulman, B.A. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
5KJX
 
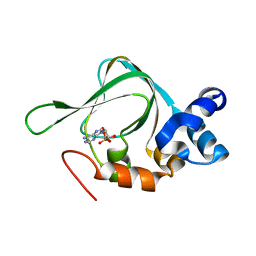 | | Co-crystal Structure of PKA RI alpha CNB-B domain with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Lorenz, R, Moon, E, Kim, J.J, Huang, G.Y, Kim, C, Herberg, F.W. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutations of PKA cyclic nucleotide-binding domains reveal novel aspects of cyclic nucleotide selectivity.
Biochem. J., 474, 2017
|
|
8PDA
 
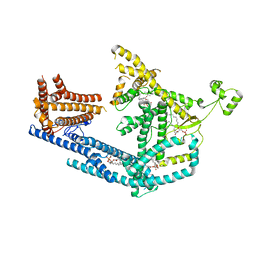 | | cryo-EM structure of Doa10 with RING domain in MSP1E3D1 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | Authors: | Botsch, J.J, Braeuning, B, Schulman, B.A. | | Deposit date: | 2023-06-12 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
5L1D
 
 | |
5L2G
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 9th Position | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(5CM)P*GP*CP*G)-3') | | Authors: | Miears, H.L, Hoppins, J.J, Gruber, D.R, Kasymov, R.D, Johnson, E.C, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-08-01 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
2O8B
 
 | | human MutSalpha (MSH2/MSH6) bound to ADP and a G T mispair | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*CP*GP*TP*GP*CP*GP*GP*TP*TP*C)-3', 5'-D(*GP*AP*AP*CP*CP*GP*CP*GP*CP*GP*CP*TP*AP*GP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
5L1M
 
 | | CASKIN2 SAM domain tandem | | Descriptor: | Caskin-2 | | Authors: | Donaldson, L.W, Kwan, J.J, Saridakis, V. | | Deposit date: | 2016-07-29 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | A new mode of SAM domain mediated oligomerization observed in the CASKIN2 neuronal scaffolding protein.
Cell Commun. Signal, 14, 2016
|
|
2O8E
 
 | | human MutSalpha (MSH2/MSH6) bound to a G T mispair, with ADP bound to MSH2 only | | Descriptor: | 5'-D(*CP*CP*TP*AP*GP*CP*CP*TP*GP*CP*GP*GP*TP*TP*C)-3', 5'-D(*GP*AP*AP*CP*CP*GP*CP*GP*GP*GP*CP*TP*AP*GP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Warren, J.J, Pohlhaus, T.J, Changela, A, Modrich, P.L, Beese, L.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the Human MutSalpha DNA Lesion Recognition Complex.
Mol.Cell, 26, 2007
|
|
5KJZ
 
 | | Co-crystal structure of PKA RI alpha CNB-B mutant (G316R/A336T) with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Lorenz, R, Moon, E, Kim, J.J, Huang, G.Y, Kim, C, Herberg, F.W. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.347 Å) | | Cite: | Mutations of PKA cyclic nucleotide-binding domains reveal novel aspects of cyclic nucleotide selectivity.
Biochem. J., 474, 2017
|
|
