1TIW
 
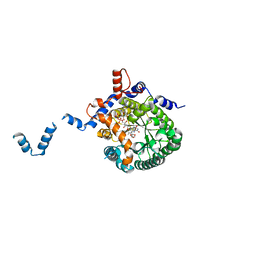 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with L-Tetrahydro-2-furoic acid | | Descriptor: | Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE, TETRAHYDROFURAN-2-CARBOXYLIC ACID | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-02 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
7SWG
 
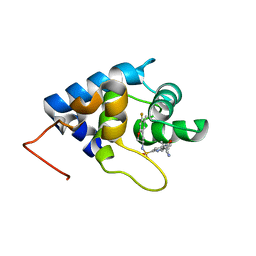 | | cTnC-TnI chimera complexed with A1 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3S)-7-fluoro-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, Troponin C, slow skeletal and cardiac muscles,Troponin I, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7SXD
 
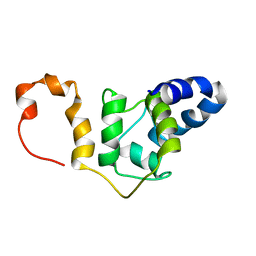 | | NMR solution structure TnC-TnI chimera | | Descriptor: | Troponin C, slow skeletal and cardiac muscles,Troponin I, cardiac muscle chimera | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-22 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
6WJ9
 
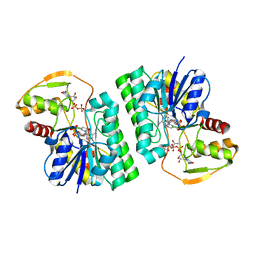 | | UDP-GlcNAc C4-epimerase mutant S121A/Y146F from Pseudomonas protegens in complex with UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Willams, R.J, Whitney, J.C, Whitfield, G.B, Robinson, H, Parsek, M.R, Nitz, M, Harrison, J.J, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
7SXC
 
 | | cTnC-TnI chimera complexed with calcium | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles,Troponin I, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-22 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
2VFQ
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666), Mutant V450A | | Descriptor: | CALCIUM ION, GLYCEROL, P22 TAILSPIKE PROTEIN,, ... | | Authors: | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | Deposit date: | 2007-11-05 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
2VFN
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666), Mutant V125A | | Descriptor: | BIFUNCTIONAL TAIL PROTEIN, CALCIUM ION, GLYCEROL, ... | | Authors: | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | Deposit date: | 2007-11-05 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
6WTW
 
 | | Structure of LaINDY crystallized in the presence of alpha-ketoglutarate and malate | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Cocco, N, Marden, J.J, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
2VFP
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666), Mutant V349L | | Descriptor: | CALCIUM ION, GLYCEROL, P22 TAILSPIKE PROTEIN, ... | | Authors: | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | Deposit date: | 2007-11-05 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
8VDD
 
 | | Crystal structure of Proinsulin C-peptide bound to HLA-DQ8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MHC class II HLA-DQ-alpha chain, ... | | Authors: | Tran, T.M, Lim, J.J, Loh, T.Y, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2023-12-14 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural basis of T cell cross-reactivity to native and spliced self-antigens presented by HLA-DQ8.
J.Biol.Chem., 300, 2024
|
|
6WU4
 
 | | Structure of the LaINDY-alpha-ketoglutarate complex | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
8TS5
 
 | | Structure of the apo FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L.L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TRT
 
 | | Structure of the EphA2 CRD bound to FabS1CE_C1, monoclinic form | | Descriptor: | CHLORIDE ION, Ephrin type-A receptor 2, S1CE variant of Fab C1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
1EI9
 
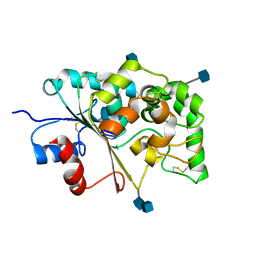 | | CRYSTAL STRUCTURE OF PALMITOYL PROTEIN THIOESTERASE 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITOYL PROTEIN THIOESTERASE 1 | | Authors: | Bellizzi III, J.J, Widom, J, Kemp, C, Lu, J.Y, Das, A.K, Hofmann, S.L, Clardy, J. | | Deposit date: | 2000-02-24 | | Release date: | 2000-04-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4LGZ
 
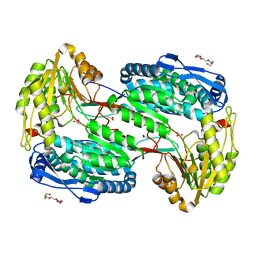 | |
6WTX
 
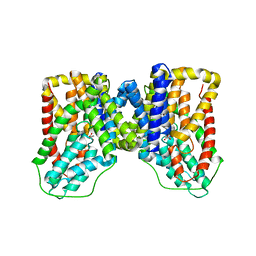 | |
8XAR
 
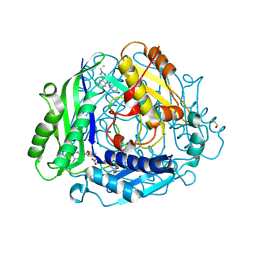 | | Structure-Based Design and Optimization of Methionine Adenosyltransferase 2A (MAT2A) Inhibitors with SAM and Compound 54 | | Descriptor: | 1,2-ETHANEDIOL, 7-chloranyl-2-ethyl-5-pyridin-3-yl-pyrazolo[3,4-c]quinolin-4-one, CHLORIDE ION, ... | | Authors: | Zheng, J.Y, Zhang, G.P, Li, J.J, Tong, S.L. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structure-Based Design and Optimization of Methionine Adenosyltransferase 2A (MAT2A) Inhibitors with High Selectivity, Brain Penetration, and In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
4L4V
 
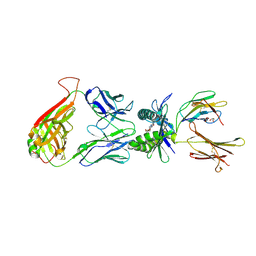 | | Structure of human MAIT TCR in complex with human MR1-RL-6-Me-7-OH | | Descriptor: | 1-deoxy-1-(7-hydroxy-6-methyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Patel, O, Kjer-Nielsen, L, Le Nours, J, Eckle, S.B.G, Birkinshaw, R.W, Beddoe, T, Corbett, A.J, Liu, L, Miles, J.J, Meehan, B, Reantragoon, R, Sandoval-Romero, M.L, Sullivan, L.C, Brooks, A.G, Chen, Z, Fairlie, D.P, McCluskey, J, Rossjohn, J. | | Deposit date: | 2013-06-09 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition of vitamin B metabolites by mucosal-associated invariant T cells.
Nat Commun, 4, 2013
|
|
1EEY
 
 | |
4LH2
 
 | |
1R6G
 
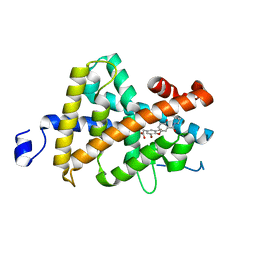 | | Crystal structure of the thyroid hormone receptor beta ligand binding domain in complex with a beta selective compound | | Descriptor: | 2-[3,5-DIBROMO-4-(4-HYDROXY-3-{HYDROXY[(2-PHENYLETHYL)AMINO]METHYL}PHENOXY)PHENYL]ETHANE-1,1-DIOL, Thyroid hormone receptor beta-1 | | Authors: | Hangeland, J.J, Dejneka, T, Friends, T.J, Devasthale, P, Mellstrom, K, Sandberg, J, Grynfarb, M, Doweyko, A.M, Sack, J.S, Einspahr, H, Farnegardh, M, Husman, B, Ljunggren, J, Koehler, K, Sheppard, C, Malm, J, Ryono, D.E. | | Deposit date: | 2003-10-15 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Thyroid receptor ligands. Part 2: Thyromimetics with improved selectivity for the thyroid hormone receptor beta.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2W55
 
 | | Crystal Structure of Xanthine Dehydrogenase (E232Q variant) from Rhodobacter capsulatus in Complex with Hypoxanthine | | Descriptor: | BARIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Doebbler, J.A, Truglio, J.J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
1R9J
 
 | | Transketolase from Leishmania mexicana | | Descriptor: | CALCIUM ION, THIAMINE DIPHOSPHATE, transketolase | | Authors: | Veitch, N.J, Mauger, D.A, Cazzulo, J.J, Lindqvist, Y, Barrett, M.P. | | Deposit date: | 2003-10-30 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Transketolase from Leishmania mexicana has a dual subcellular localization.
Biochem.J., 382, 2004
|
|
5OL9
 
 | | Structure of human mitochondrial transcription elongation factor (TEFM) N-terminal domain | | Descriptor: | ACETATE ION, Transcription elongation factor, mitochondrial | | Authors: | Hillen, H.S, Parshin, A.V, Agaronyan, K, Morozov, Y, Graber, J.J, Chernev, A, Schwinghammer, K, Urlaub, H, Anikin, M, Cramer, P, Temiakov, D. | | Deposit date: | 2017-07-27 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Mechanism of Transcription Anti-termination in Human Mitochondria.
Cell, 171, 2017
|
|
2W54
 
 | | Crystal Structure of Xanthine Dehydrogenase from Rhodobacter capsulatus in Complex with Bound Inhibitor Pterin-6-aldehyde | | Descriptor: | 6-HYDROXYMETHYLPTERIN, BARIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Doebbler, J.A, Truglio, J.J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
