8EYC
 
 | |
8FN4
 
 | | Cryo-EM structure of RNase-treated RESC-A in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), RNA-editing substrate-binding complex protein 3 (RESC3), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FN6
 
 | | Cryo-EM structure of RNase-untreated RESC-A in trypanosomal RNA editing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8T8K
 
 | |
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNF
 
 | | Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8T8L
 
 | |
4JNZ
 
 | |
8FNI
 
 | | Cryo-EM structure of RNase-treated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNK
 
 | | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
6WU1
 
 | | Structure of apo LaINDY | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N.C, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
1SIR
 
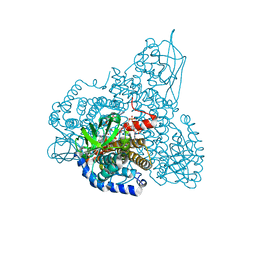 | | The Crystal Structure and Mechanism of Human Glutaryl-CoA Dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, S-4-NITROBUTYRYL-COA | | Authors: | Wang, M, Fu, Z, Paschke, R, Goodman, S.L, Frerman, F.E, Kim, J.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Human Glutaryl-CoA Dehydrogenase with and without an Alternate Substrate: Structural Bases of Dehydrogenation and Decarboxylation Reactions
Biochemistry, 43, 2004
|
|
8T9X
 
 | |
8TA7
 
 | |
8T9E
 
 | |
8TTJ
 
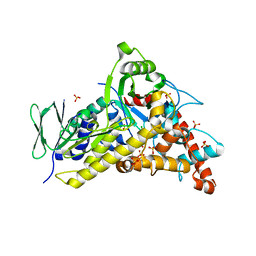 | | Tryptophan-6-halogenase BorH complexed with 6-chlorotryptophan | | Descriptor: | 6-CHLORO-L-TRYPTOPHAN, SULFATE ION, Tryptophan 6-halogenase | | Authors: | Lingkon, K, Bellizzi, J.J. | | Deposit date: | 2023-08-14 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic and thermodynamic evidence of negative cooperativity of flavin and tryptophan binding in the flavin-dependent halogenases AbeH and BorH.
Biorxiv, 2023
|
|
6X3Z
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Kim, J.J, Gharpure, A, Teng, J, Zhuang, Y, Howard, R.J, Zhu, S, Noviello, C.M, Walsh, R.M, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Shared structural mechanisms of general anaesthetics and benzodiazepines.
Nature, 585, 2020
|
|
8T9C
 
 | |
1SFP
 
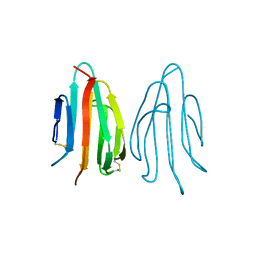 | | CRYSTAL STRUCTURE OF ACIDIC SEMINAL FLUID PROTEIN (ASFP) AT 1.9 A RESOLUTION: A BOVINE POLYPEPTIDE FROM THE SPERMADHESIN FAMILY | | Descriptor: | ASFP | | Authors: | Romao, M.J, Kolln, I, Dias, J.M, Carvalho, A.L, Romero, A, Varela, P.F, Sanz, L, Topfer-Petersen, E, Calvete, J.J. | | Deposit date: | 1997-06-24 | | Release date: | 1998-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of two spermadhesins reveal the CUB domain fold.
Nat.Struct.Biol., 4, 1997
|
|
8TJE
 
 | |
8TTI
 
 | | Trp-6-Halogenase BorH complexed with FAD and Trp | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TRYPTOPHAN, ... | | Authors: | Lingkon, K, Bellizzi, J.J. | | Deposit date: | 2023-08-14 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic and thermodynamic evidence of negative cooperativity of flavin and tryptophan binding in the flavin-dependent halogenases AbeH and BorH.
Biorxiv, 2023
|
|
8TTK
 
 | | Tryptophan-6-halogenase BorH apo structure | | Descriptor: | SULFATE ION, Tryptophan 6-halogenase | | Authors: | Lingkon, K, Bellizzi, J.J. | | Deposit date: | 2023-08-14 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic and thermodynamic evidence of negative cooperativity of flavin and tryptophan binding in the flavin-dependent halogenases AbeH and BorH.
Biorxiv, 2023
|
|
6X3V
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus etomidate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Etomidate, ... | | Authors: | Kim, J.J, Gharpure, A, Teng, J, Zhuang, Y, Howard, R.J, Zhu, S, Noviello, C.M, Walsh, R.M, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Shared structural mechanisms of general anaesthetics and benzodiazepines.
Nature, 585, 2020
|
|
6X0K
 
 | |
6X3T
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus propofol | | Descriptor: | 2,6-BIS(1-METHYLETHYL)PHENOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, J.J, Gharpure, A, Teng, J, Zhuang, Y, Howard, R.J, Zhu, S, Noviello, C.M, Walsh, R.M, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Shared structural mechanisms of general anaesthetics and benzodiazepines.
Nature, 585, 2020
|
|
