4BIP
 
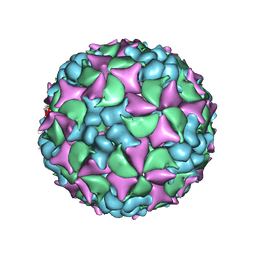 | | Homology model of coxsackievirus A7 (CAV7) full capsid proteins. | | Descriptor: | VP1, VP2, VP3 | | Authors: | Seitsonen, J.J.T, Shakeel, S, Susi, P, Pandurangan, A.P, Sinkovits, R.S, Hyvonen, H, Laurinmaki, P, Yla-Pelto, J, Topf, M, Hyypia, T, Butcher, S.J. | | Deposit date: | 2013-04-12 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.23 Å) | | Cite: | Combined Approaches to Flexible Fitting and Assessment in Virus Capsids Undergoing Conformational Change.
J.Struct.Biol., 185, 2014
|
|
3UO9
 
 | | Crystal Structure of Human GAC in Complex with Glutamate and BPTES | | Descriptor: | GLYCEROL, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | DeLaBarre, B, Gross, S, Cheng, F, Gao, Y, Jha, A, Jiang, F, Song, J.J, Wei, W, Hurov, J.B. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Full-length human glutaminase in complex with an allosteric inhibitor.
Biochemistry, 50, 2011
|
|
2V3V
 
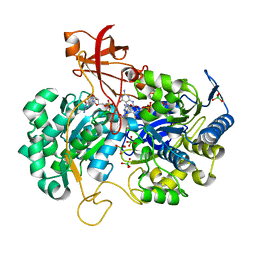 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-22 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
4QJ4
 
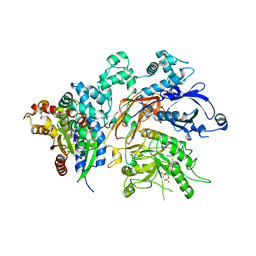 | | Structure of a fragment of human phospholipase C-beta3 delta472-569, bound to IP3 and in complex with Galphaq | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Lyon, A.M, Tesmer, J.J.G. | | Deposit date: | 2014-06-03 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular mechanisms of phospholipase C beta 3 autoinhibition.
Structure, 22, 2014
|
|
6D8Z
 
 | |
8XUV
 
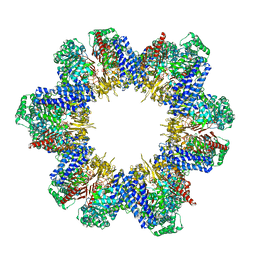 | | Cryo-EM structure of tomato NRC2 filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, NRC2 | | Authors: | Sun, Y, Ma, S.C, Chai, J.J. | | Deposit date: | 2024-01-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR.
Nature, 632, 2024
|
|
8XUO
 
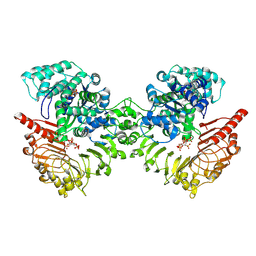 | | Cryo-EM structure of tomato NRC2 dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, NRC2 | | Authors: | Sun, Y, Ma, S.C, Chai, J.J. | | Deposit date: | 2024-01-13 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR.
Nature, 632, 2024
|
|
8XUQ
 
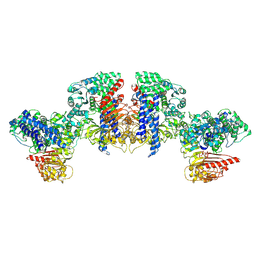 | | Cryo-EM structure of tomato NRC2 tetramer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, NRC2 | | Authors: | Sun, Y, Ma, S.C, Chai, J.J. | | Deposit date: | 2024-01-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR.
Nature, 632, 2024
|
|
4PNI
 
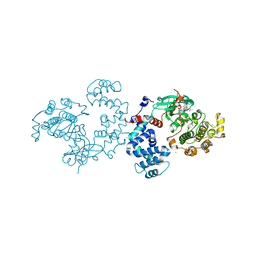 | | Bovine G protein-coupled receptor kinase 1 in complex with GSK2163632A | | Descriptor: | 3-[(2-{[1-(N,N-dimethylglycyl)-6-methoxy-4,4-dimethyl-1,2,3,4-tetrahydroquinolin-7-yl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]thiophene-2-carboxamide, CHLORIDE ION, Rhodopsin kinase | | Authors: | Homan, K.T, Tesmer, J.J.G. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and structure-function analysis of subfamily selective g protein-coupled receptor kinase inhibitors.
Acs Chem.Biol., 10, 2015
|
|
4BEZ
 
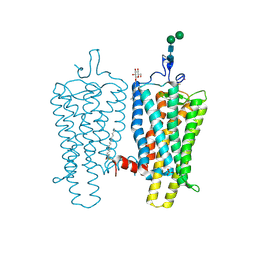 | | Night blindness causing G90D rhodopsin in the active conformation | | Descriptor: | ACETATE ION, PALMITIC ACID, RHODOPSIN, ... | | Authors: | Singhal, A, Ostermaier, M.K, Vishnivetskiy, S.A, Panneels, V, Homan, K.T, Tesmer, J.J.G, Veprintsev, D, Deupi, X, Gurevich, V.V, Schertler, G.F.X, Standfuss, J. | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insights Into Congenital Stationary Night Blindness Based on the Structure of G90D Rhodopsin.
Embo Rep., 14, 2013
|
|
4BIQ
 
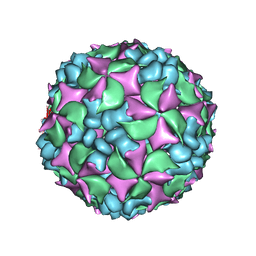 | | Homology model of coxsackievirus A7 (CAV7) empty capsid proteins. | | Descriptor: | VP1, VP2, VP3 | | Authors: | Seitsonen, J.J.T, Shakeel, S, Susi, P, Pandurangan, A.P, Sinkovits, R.S, Hyvonen, H, Laurinmaki, P, Yla-Pelto, J, Topf, M, Hyypia, T, Butcher, S.J. | | Deposit date: | 2013-04-12 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.09 Å) | | Cite: | Combined Approaches to Flexible Fitting and Assessment in Virus Capsids Undergoing Conformational Change.
J.Struct.Biol., 185, 2014
|
|
6Y0O
 
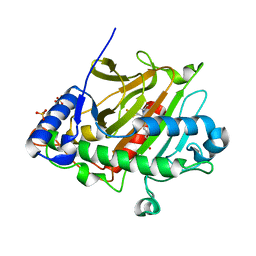 | | isopenicillin N synthase in complex with ACV and Fe under anaerobic environment using FT-SSX methods | | Descriptor: | Anaerobic Fixed Target Structure of Isopenicillin N synthase in complex with Fe and ACV, FE (III) ION, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Rabe, P, Beale, J.H, Lang, P.A, Dirr, A.S, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anaerobic fixed-target serial crystallography.
Iucrj, 7, 2020
|
|
5URI
 
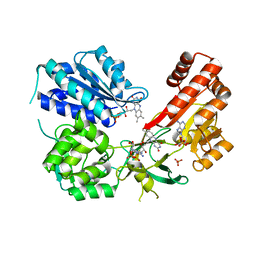 | | Rat CYPOR/D632A with 2'-AMP | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Kinetic Studies of Asp632 Mutants and Fully Reduced NADPH-Cytochrome P450 Oxidoreductase Define the Role of Asp632 Loop Dynamics in the Control of NADPH Binding and Hydride Transfer.
Biochemistry, 57, 2018
|
|
8FAM
 
 | | Edited Octopus bimaculoides Synaptotagmin 1 C2A (I248V) at room temperature | | Descriptor: | Synaptotagmin | | Authors: | McNeme, S, Dominguez, M.J, Birk, M.A, Rosenthal, J.J, Sutton, R.B. | | Deposit date: | 2022-11-28 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Temperature-dependent RNA editing in octopus extensively recodes the neural proteome.
Cell, 186, 2023
|
|
5URE
 
 | | Wild type rat CYPOR bound with NADP+ - reduced form | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Studies of Asp632 Mutants and Fully Reduced NADPH-Cytochrome P450 Oxidoreductase Define the Role of Asp632 Loop Dynamics in the Control of NADPH Binding and Hydride Transfer.
Biochemistry, 57, 2018
|
|
8WJQ
 
 | | Cryo-EM structure of URAT1(R477S)-Urate complex | | Descriptor: | Solute carrier family 22 member 12, URIC ACID | | Authors: | Qian, H.W, He, J.J. | | Deposit date: | 2023-09-26 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the transport and substrate selection of human urate transporter 1.
Cell Rep, 43, 2024
|
|
8WJH
 
 | | Cryo-EM structure of OAT4 | | Descriptor: | CHLORIDE ION, Solute carrier family 22 member 11 | | Authors: | Qian, H.W, He, J.J. | | Deposit date: | 2023-09-26 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the transport and substrate selection of human urate transporter 1.
Cell Rep, 43, 2024
|
|
8WJG
 
 | | Cryo-EM structure of URAT1(R477S) | | Descriptor: | Solute carrier family 22 member 12 | | Authors: | Qian, H.W, He, J.J. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the transport and substrate selection of human urate transporter 1.
Cell Rep, 43, 2024
|
|
4QJ3
 
 | | Structure of a fragment of human phospholipase C-beta3 delta472-559, in complex with Galphaq | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lyon, A.M, Tesmer, J.J.G. | | Deposit date: | 2014-06-03 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular mechanisms of phospholipase C beta 3 autoinhibition.
Structure, 22, 2014
|
|
2V45
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-27 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
5VT4
 
 | | Sco GlgEI-V279S in complex with a pyrolidene-based methyl-phosphonate compound | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, {[(2R,3R,4R,5R)-3-(alpha-D-glucopyranosyloxy)-4-hydroxy-2,5-bis(hydroxymethyl)pyrrolidin-1-yl]methyl}phosphonic acid | | Authors: | Petit, C, Ronning, D.R, Lindenberger, J.J. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Zwitterionic pyrrolidene-phosphonates: inhibitors of the glycoside hydrolase-like phosphorylase Streptomyces coelicolor GlgEI-V279S.
Org. Biomol. Chem., 15, 2017
|
|
4PNK
 
 | | G protein-coupled receptor kinase 2 in complex with GSK180736A | | Descriptor: | (4S)-4-(4-fluorophenyl)-N-(2H-indazol-5-yl)-6-methyl-2-oxo-1,2,3,4-tetrahydropyrimidine-5-carboxamide, Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Homan, K.T, Larimore, K.M, Elkins, J, Knapp, S, Tesmer, J.J.G. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Identification and structure-function analysis of subfamily selective g protein-coupled receptor kinase inhibitors.
Acs Chem.Biol., 10, 2015
|
|
4QJ5
 
 | | Structure of a fragment of human phospholipase C-beta3 delta472-581, bound to IP3 and in complex with Galphaq | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Lyon, A.M, Tesmer, J.J.G. | | Deposit date: | 2014-06-03 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Molecular mechanisms of phospholipase C beta 3 autoinhibition.
Structure, 22, 2014
|
|
4M6B
 
 | | Crystal structure of yeast Swr1-Z domain in complex with H2A.Z-H2B dimer | | Descriptor: | Chimera protein of Histone H2B.1 and Histone H2A.Z, Helicase SWR1 | | Authors: | Hong, J.J, Feng, H.Q, Wang, F, Ranjan, A, Chen, J.H, Jiang, J.S, Girlando, R, Xiao, T.S, Wu, C, Bai, Y.W. | | Deposit date: | 2013-08-09 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Catalytic Subunit of the SWR1 Remodeler Is a Histone Chaperone for the H2A.Z-H2B Dimer.
Mol.Cell, 53, 2014
|
|
2KKD
 
 | | NMR Structure of Ni Substitued Desulfovibrio vulgaris Rubredoxin | | Descriptor: | NICKEL (II) ION, Rubredoxin | | Authors: | Nunes, S.G, Volkman, B.F, Moura, J.J.G, Moura, I, Macedo, A.L, Markley, J.L, Duarte, I.C. | | Deposit date: | 2009-06-18 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR structural study of nickel-substituted rubredoxin
J.Biol.Inorg.Chem., 15, 2010
|
|
