2AEW
 
 | |
6ZAM
 
 | | Fluorine labeled IPNS S55C in complex with Fe and ACV under anaerobic conditions. | | Descriptor: | 1,1,1-tris(fluoranyl)propan-2-one, 1,1,1-tris(fluoranyl)propane-2,2-diol, FE (III) ION, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-06-05 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
6ZAL
 
 | | Room temperature XFEL Isopenicillin N synthase structure in complex with Fe and ACV after exposure to dioxygen for 500ms without glycerol. | | Descriptor: | FE (II) ION, Isopenicillin N synthase, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-06-05 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
6ZAP
 
 | | Isopenicillin N synthase in complex with Fe, O2 and ACV under cryo conditions. | | Descriptor: | FE (III) ION, Isopenicillin N synthase, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-06-05 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
6ZAF
 
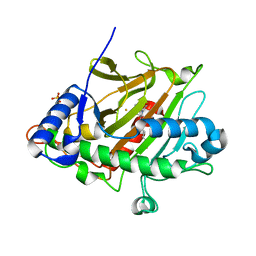 | | Room temperature XFEL Isopenicillin N synthase structure in complex with Fe and ACV after exposure to dioxygen for 400ms. | | Descriptor: | FE (II) ION, Isopenicillin N synthase, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-06-05 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4001 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
6ZAJ
 
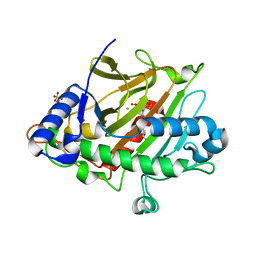 | | Room temperature XFEL Isopenicillin N synthase structure in complex with Fe, O2 and ACV after exposure to dioxygen for 3000ms. | | Descriptor: | FE (II) ION, Isopenicillin N synthase, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-06-05 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5301 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
2AAL
 
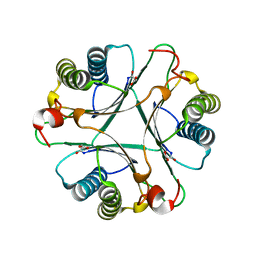 | | Crystal Structures of the Wild-type, Mutant-P1A and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities | | Descriptor: | MALONATE ION, Malonate semialdehyde decarboxylase | | Authors: | Almrud, J.J, Poelarends, G.J, Johnson Jr, W.H, Serrano, H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2005-07-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of the Wild-Type, P1A Mutant, and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities
Biochemistry, 44, 2005
|
|
6ZAK
 
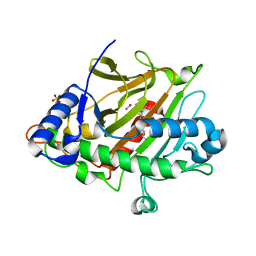 | | Room temperature XFEL Isopenicillin N synthase structure in complex with the Fe(IV)=O mimic VO and ACV. | | Descriptor: | Isopenicillin N synthase, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, SULFATE ION, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-06-05 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Room temperature XFEL Isopenicillin N synthase structure in complex with the Fe(IV)=O mimic VO and ACV.
To Be Published
|
|
2AAJ
 
 | | Crystal Structures of the Wild-type, Mutant-P1A and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities | | Descriptor: | Malonate Semialdehyde Decarboxylase | | Authors: | Almrud, J.J, Poelarends, G.J, Johnson Jr, W.H, Serrano, H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2005-07-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal Structures of the Wild-Type, P1A Mutant, and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities
Biochemistry, 44, 2005
|
|
6ZS5
 
 | | 3.5 A cryo-EM structure of human uromodulin filament core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
8HDG
 
 | | Small peptide enhances the binding of nutline-3a to MdmX | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, Uncharacterized protein DKFZp686B01123 | | Authors: | Cheng, X.Y, Huang, Y, Wei, Q.Y, Huang, J.J, Peng, Y.W, Su, Z.D. | | Deposit date: | 2022-11-04 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Small peptide enhances the binding of nutline-3a to MdmX
To Be Published
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6ZPH
 
 | | Kinesin binding protein complexed with Kif15 motor domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KIF-binding protein, Kinesin-like protein KIF15, ... | | Authors: | Atherton, J, Hummel, J.J.A, Olieric, N, Locke, J, Pena, A, Rosenfeld, S.S, Steinmetz, M.O, Hoogenraad, C.C, Moores, C.A. | | Deposit date: | 2020-07-08 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | The mechanism of kinesin inhibition by kinesin-binding protein.
Elife, 9, 2020
|
|
2B35
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (InhA) inhibited by triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Sullivan, T.J, Truglio, J.J, Novichenok, P, Stratton, C, Zhang, X, Kaur, T, Johnson, F, Boyne, M.S, Amin, A. | | Deposit date: | 2005-09-19 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High Affinity InhA Inhibitors with Activity against Drug-Resistant Strains
of Mycobacterium tuberculosis
ACS Chem.Biol., 1, 2006
|
|
2B4F
 
 | | Structure Of A Cold-Adapted Family 8 Xylanase in complex with substrate | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-xylanase | | Authors: | De Vos, D, Collins, T, Savvides, S.N, Feller, G, Van Beeumen, J.J. | | Deposit date: | 2005-09-23 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Oligosaccharide binding in family 8 glycosidases: crystal structures of active-site mutants of the beta-1,4-xylanase pXyl from Pseudoaltermonas haloplanktis TAH3a in complex with substrate and product.
Biochemistry, 45, 2006
|
|
5Y7A
 
 | |
4GGJ
 
 | | Crystal structure of Zucchini from mouse (mZuc / PLD6 / MitoPLD) | | Descriptor: | Mitochondrial cardiolipin hydrolase, ZINC ION | | Authors: | Ipsaro, J.J, Haase, A.D, Hannon, G.J, Joshua-Tor, L. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis.
Nature, 491, 2012
|
|
4FUM
 
 | |
2AKQ
 
 | | The structure of bovine B-lactoglobulin A in crystals grown at very low ionic strength | | Descriptor: | Beta-lactoglobulin variant A | | Authors: | Adams, J.J, Anderson, B.F, Norris, G.E, Creamer, L.K, Jameson, G.B. | | Deposit date: | 2005-08-03 | | Release date: | 2005-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bovine beta-lactoglobulin (variant A) at very low ionic strength
J.Struct.Biol., 154, 2006
|
|
2AXG
 
 | | The Immunogenicity of a Viral Cytotoxic T Cell Epitope is controlled by its MHC-bound Conformation | | Descriptor: | 10-mer peptide from BZLF1 trans-activator protein, ACETIC ACID, Beta-2-microglobulin, ... | | Authors: | Tynan, F.E, Elhassen, D, Purcell, A.W, Burrows, J.M, Borg, N.A, Miles, J.J, Williamson, N.A, Green, K.J, Tellam, J, Kjer-Nielsen, L, McCluskey, J, Rossjohn, J, Burrows, S.R. | | Deposit date: | 2005-09-05 | | Release date: | 2005-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The immunogenicity of a viral cytotoxic T cell epitope is controlled by its MHC-bound conformation
J.Exp.Med., 202, 2005
|
|
4G85
 
 | | Crystal structure of human HisRS | | Descriptor: | Histidine-tRNA ligase, cytoplasmic | | Authors: | Wei, Z, Wu, J, Zhou, J.J, Yang, X.-L, Zhang, M, Schimmel, P. | | Deposit date: | 2012-07-21 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Internally Deleted Human tRNA Synthetase Suggests Evolutionary Pressure for Repurposing.
Structure, 20, 2012
|
|
2B0D
 
 | | EcoRV Restriction Endonuclease/GAATTC/Ca2+ | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*AP*TP*TP*CP*TP*T)-3', CALCIUM ION, Type II restriction enzyme EcoRV | | Authors: | Hiller, D.A, Rodriguez, A.M, Perona, J.J. | | Deposit date: | 2005-09-13 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-cognate Enzyme-DNA Complex: Structural and Kinetic Analysis of EcoRV Endonuclease Bound to the EcoRI Recognition Site GAATTC
J.Mol.Biol., 354, 2005
|
|
2B36
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (InhA) inhibited by 5-pentyl-2-phenoxyphenol | | Descriptor: | 5-PENTYL-2-PHENOXYPHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sullivan, T.J, Truglio, J.J, Novichenok, P, Stratton, C, Zhang, X, Kaur, T, Johnson, F, Boyne, M.S, Amin, A. | | Deposit date: | 2005-09-19 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High Affinity InhA Inhibitors with Activity against Drug-Resistant Strains
of Mycobacterium tuberculosis
ACS Chem.Biol., 1, 2006
|
|
5Y77
 
 | |
7DYS
 
 | |
