8TD9
 
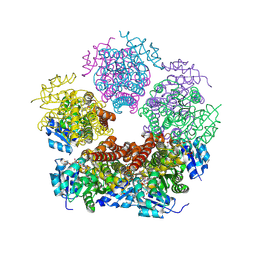 | | Structure of PYCR1 complexed with NADH and L-pipecolic acid | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD3
 
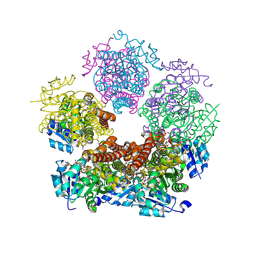 | |
8TDB
 
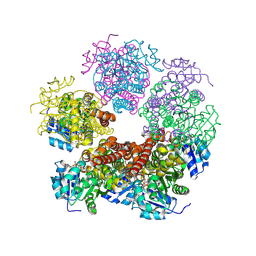 | | Structure of PYCR1 complexed with NADH and 1-hydroxyethane-1-sulfonate | | Descriptor: | (1R)-1-hydroxyethane-1-sulfonic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD6
 
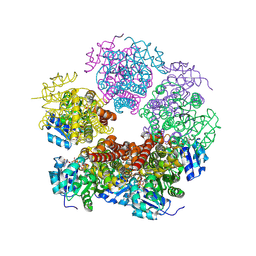 | | Structure of PYCR1 complexed with NADH and 2-(Methylthio)acetic acid | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD4
 
 | | Structure of PYCR1 complexed with NADH and 1,3-Dithiolane-2-carboxylic acid | | Descriptor: | 1,3-dithiolane-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD2
 
 | | Structure of PYCR1 complexed with NADH and cyclobutane-1,1-dicarboxylic acid | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD7
 
 | | Structure of PYCR1 complexed with 2S-hydroxy-3-methylbutyric acid | | Descriptor: | (2S)-2-hydroxy-3-methylbutanoic acid, DI(HYDROXYETHYL)ETHER, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TDD
 
 | | Structure of PYCR1 complexed with NADH and 2-(furan-2-yl)acetic acid | | Descriptor: | (furan-2-yl)acetic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TD8
 
 | | Structure of PYCR1 complexed with NADH and 2S-Hydroxy-3,3-dimethylbutyric acid | | Descriptor: | (2S)-2-hydroxy-3,3-dimethylbutanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TRV
 
 | | Structure of the EphA2 LBDCRD bound to FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ephrin type-A receptor 2, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Synthetic Antibodies targeting EPHA2 Induce Diverse Signaling-Competent Clusters with Differential Activation
To be Published
|
|
8TD5
 
 | | Structure of PYCR1 complexed with NADH and Tetrahydrothiophene-2-carboxylic acid | | Descriptor: | (2R)-thiolane-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8TRL
 
 | | T cell recognition of citrullinated alpha-enolase peptide presented by HLA-DR4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lim, J.J, Loh, T.J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2023-08-09 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular basis underlying T cell specificity towards citrullinated epitopes presented by HLA-DR4.
Nat Commun, 15, 2024
|
|
8TV2
 
 | | Structure of apo FabS1C_L1 | | Descriptor: | CHLORIDE ION, S1C variant of Fab_L1 heavy chain, S1C variant of Fab_L1 light chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-17 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthetic Antibodies targeting EPHA2 Induce Diverse Signaling-Competent Clusters with Differential Activation
To be published
|
|
8TRQ
 
 | | T cell recognition of citrullinated vimentin peptide presented by HLA-DR4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A07 TCR alpha chain, A07 TCR beta chain, ... | | Authors: | Loh, T.J, Lim, J.J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2023-08-10 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The molecular basis underlying T cell specificity towards citrullinated epitopes presented by HLA-DR4.
Nat Commun, 15, 2024
|
|
8TRR
 
 | | T cell recognition of citrullinated vimentin peptide presented by HLA-DR4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Loh, T.J, Lim, J.J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2023-08-10 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The molecular basis underlying T cell specificity towards citrullinated epitopes presented by HLA-DR4.
Nat Commun, 15, 2024
|
|
6UJU
 
 | | Structure of the HIV-1 gp41 transmembrane domain and cytoplasmic tail (LLP2) | | Descriptor: | Envelope glycoprotein GP41 | | Authors: | Piai, A, Fu, Q, Cai, Y, Ghantous, F, Xiao, T, Shaik, M.M, Peng, H, Rits-Volloch, S, Liu, Z, Chen, W, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein.
Nat Commun, 11, 2020
|
|
2R0N
 
 | | The effect of a Glu370Asp mutation in Glutaryl-CoA Dehydrogenase on Proton Transfer to the Dienolate Intermediate | | Descriptor: | 3-thiaglutaryl-CoA, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase | | Authors: | Rao, K.S, Albro, M, Fu, Z, Narayanan, B, Baddam, S, Lee, H.J, Kim, J.J, Frerman, F.E. | | Deposit date: | 2007-08-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The effect of a Glu370Asp mutation in glutaryl-CoA dehydrogenase on proton transfer to the dienolate intermediate.
Biochemistry, 46, 2007
|
|
4IMK
 
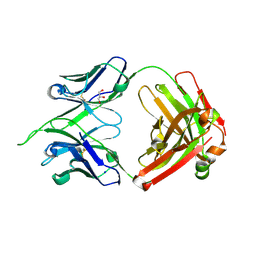 | | Uncrossed Fab binding to human Angiopoietin 2 | | Descriptor: | GLYCEROL, Heavy Chain, Light Chain, ... | | Authors: | Fenn, S, Schiller, C, Griese, J.J, Hopfner, K.-P, Kettenberger, H. | | Deposit date: | 2013-01-03 | | Release date: | 2013-04-17 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Crystal Structure of an Anti-Ang2 CrossFab Demonstrates Complete Structural and Functional Integrity of the Variable Domain.
Plos One, 8, 2013
|
|
1SX5
 
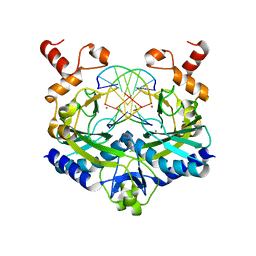 | |
1T24
 
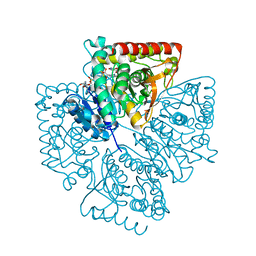 | | Plasmodium falciparum lactate dehydrogenase complexed with NAD+ and 4-hydroxy-1,2,5-oxadiazole-3-carboxylic acid | | Descriptor: | 4-HYDROXY-1,2,5-OXADIAZOLE-3-CARBOXYLIC ACID, GLYCEROL, L-lactate dehydrogenase, ... | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
6UXI
 
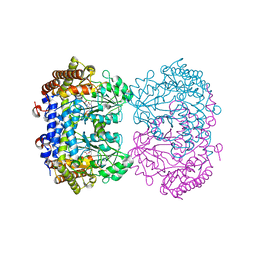 | | Structure of serine hydroxymethyltransferase 8 from Glycine max cultivar Essex complexed with PLP-Glycine | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], Serine hydroxymethyltransferase | | Authors: | Korasick, D.A, Tanner, J.J, Beamer, L.J. | | Deposit date: | 2019-11-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Impaired folate binding of serine hydroxymethyltransferase 8 from soybean underlies resistance to the soybean cyst nematode.
J.Biol.Chem., 295, 2020
|
|
8DQ2
 
 | | X-ray crystal structure of Hansschlegelia quercus lanmodulin (LanM) with lanthanum (III) bound at pH 7 | | Descriptor: | CITRIC ACID, EF-hand domain-containing protein, LANTHANUM (III) ION, ... | | Authors: | Jung, J.J, Lin, C.-Y, Boal, A.K. | | Deposit date: | 2022-07-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enhanced rare-earth separation with a metal-sensitive lanmodulin dimer.
Nature, 618, 2023
|
|
1SX8
 
 | | EcoRV bound to cognate DNA and Mn2+ | | Descriptor: | 5'-D(*C*AP*AP*GP*AP*TP*AP*TP*CP*TP*T)-3', MANGANESE (II) ION, Type II restriction enzyme EcoRV | | Authors: | Horton, N.C, Perona, J.J. | | Deposit date: | 2004-03-30 | | Release date: | 2004-04-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | DNA Cleavage by EcoRV Endonuclease: Two Metal Ions in Three Metal Ion Binding Sites
Biochemistry, 43, 2004
|
|
6UXK
 
 | |
6V1A
 
 | | immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta 74cit69-81, GLYCEROL, ... | | Authors: | Lim, J.J, Rossjohn, J. | | Deposit date: | 2019-11-20 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR.
Sci Immunol, 6, 2021
|
|
