3ZO2
 
 | | The Synthesis and Evaluation of Diazaspirocyclic Protein Kinase Inhibitors | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-(2,9-Diazaspiro[5.5]undecan-9-yl)-9H-purine, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, ... | | Authors: | Allen, C.E, Chow, C.L, Caldwell, J.J, Westwood, I.M, van Montfort, R.L, Collins, I. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Synthesis and evaluation of heteroaryl substituted diazaspirocycles as scaffolds to probe the ATP-binding site of protein kinases.
Bioorg. Med. Chem., 21, 2013
|
|
1RJL
 
 | | Structure of the complex between OspB-CT and bactericidal Fab-H6831 | | Descriptor: | Fab H6831 H-chain, Fab H6831 L-chain, Outer surface protein B | | Authors: | Becker, M, Bunikis, J, Lade, B.D, Dunn, J.J, Barbour, A.G, Lawson, C.L. | | Deposit date: | 2003-11-19 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Investigation of Borrelia burgdorferi OspB, a BactericidalFab Target.
J.Biol.Chem., 280, 2005
|
|
7TSR
 
 | | Room temperature rsEospa Cis-state structure at pH 8.4 | | Descriptor: | Cis-state rsEospa | | Authors: | Baxter, J.M, van Thor, J.J. | | Deposit date: | 2022-01-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Observation of Cation Chromophore Photoisomerization of a Fluorescent Protein Using Millisecond Synchrotron Serial Crystallography and Infrared Vibrational and Visible Spectroscopy.
J.Phys.Chem.B, 126, 2022
|
|
3ZZE
 
 | | Crystal structure of C-MET kinase domain in complex with N'-((3Z)-4- chloro-7-methyl-2-oxo-1,2-dihydro-3H-indol-3-ylidene)-2-(4- hydroxyphenyl)propanohydrazide | | Descriptor: | (2S)-N'-[(3R)-4-chloro-7-methyl-2-oxo-2,3-dihydro-1H-indol-3-yl]-2-(4-hydroxyphenyl)propanehydrazide, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Deng, Y, Ryan, K, Cui, J.J. | | Deposit date: | 2011-08-31 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
8ASA
 
 | | Crystal structure of AO75L | | Descriptor: | 1,2-ETHANEDIOL, BICINE, CALCIUM ION, ... | | Authors: | Laugeri, M.E, Speciale, I, Gimeno, A, Lin, S, Poveda, A, Lowary, T, Van Etten, J.L, Barbero, J.J, De Castro, C, Tonetti, M, Rojas, A.L. | | Deposit date: | 2022-08-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of AO75L
To Be Published
|
|
7TSU
 
 | | Room temperature rsEospa Cis-state structure at pH 5.5 | | Descriptor: | Cis-state rsEospa | | Authors: | Baxter, J.M, van Thor, J.J. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Observation of Cation Chromophore Photoisomerization of a Fluorescent Protein Using Millisecond Synchrotron Serial Crystallography and Infrared Vibrational and Visible Spectroscopy.
J.Phys.Chem.B, 126, 2022
|
|
7TSS
 
 | | Room temperature rsEospa Trans-state structure at pH 8.4 | | Descriptor: | Trans-state rsEospa | | Authors: | Baxter, J.M, van Thor, J.J. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Observation of Cation Chromophore Photoisomerization of a Fluorescent Protein Using Millisecond Synchrotron Serial Crystallography and Infrared Vibrational and Visible Spectroscopy.
J.Phys.Chem.B, 126, 2022
|
|
1U04
 
 | |
3ZSO
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based design | | Descriptor: | 5-{[(2-{[bis(4-methoxyphenyl)methyl]carbamoyl}benzyl)(prop-2-en-1-yl)amino]methyl}-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, INTEGRASE, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Deadman, J.J, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L. | | Deposit date: | 2011-06-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
7TSV
 
 | | Room temperature rsEospa Trans-state structure at pH 5.5 | | Descriptor: | Trans-state rsEospa | | Authors: | Baxter, J.M, van Thor, J.J. | | Deposit date: | 2022-01-31 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Observation of Cation Chromophore Photoisomerization of a Fluorescent Protein Using Millisecond Synchrotron Serial Crystallography and Infrared Vibrational and Visible Spectroscopy.
J.Phys.Chem.B, 126, 2022
|
|
1FF9
 
 | | APO SACCHAROPINE REDUCTASE | | Descriptor: | SACCHAROPINE REDUCTASE, SULFATE ION | | Authors: | Johansson, E, Steffens, J.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 2000-07-25 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of saccharopine reductase from Magnaporthe grisea, an enzyme of the alpha-aminoadipate pathway of lysine biosynthesis.
Structure Fold.Des., 8, 2000
|
|
3ZO4
 
 | | The Synthesis and Evaluation of Diazaspirocyclic Protein Kinase Inhibitors | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-(4-PHENYL-1,9-DIAZASPIRO[5.5]UNDECAN-9-YL)-9H-PURINE, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, ... | | Authors: | Allen, C.E, Chow, C.L, Caldwell, J.J, Westwood, I.M, van Montfort, R.L, Collins, I. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Synthesis and evaluation of heteroaryl substituted diazaspirocycles as scaffolds to probe the ATP-binding site of protein kinases.
Bioorg. Med. Chem., 21, 2013
|
|
4JZT
 
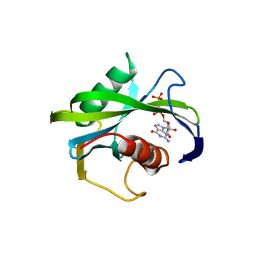 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH (E68A mutant) bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, dGTP pyrophosphohydrolase | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3M48
 
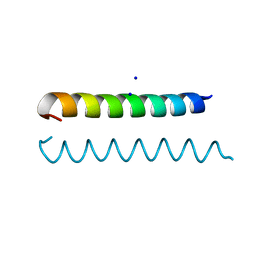 | | GCN4 Leucine Zipper Peptide Mutant | | Descriptor: | General control protein GCN4, SODIUM ION | | Authors: | Du, S, Kettering, R.D, Alvarado, J.J, Tortajada, A, Yeh, J.I. | | Deposit date: | 2010-03-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | GCN4 Leucine Zipper Peptide Mutant
To be Published
|
|
7STT
 
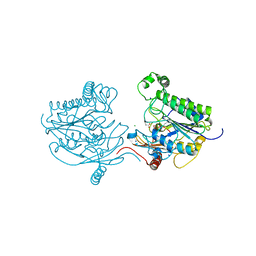 | | Crystal structure of sulfatase from Pedobacter yulinensis | | Descriptor: | CALCIUM ION, CHLORIDE ION, MALONATE ION, ... | | Authors: | O'Malley, A, Schlachter, C.R, Grimes, L.L, Tomashek, J.J, Lee, A.L, Chruszcz, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis .
Molecules, 27, 2021
|
|
3MGC
 
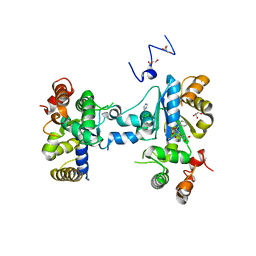 | | Teg12 Apo | | Descriptor: | GLYCEROL, IMIDAZOLE, N-L-ALPHA-ASPARTYL L-PHENYLALANINE 1-METHYL ESTER, ... | | Authors: | Bick, M.J, Banik, J.J, Darst, S.A, Brady, S.F. | | Deposit date: | 2010-04-05 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structures of the glycopeptide sulfotransferase Teg12 in a complex with the teicoplanin aglycone.
Biochemistry, 49, 2010
|
|
7STV
 
 | | Crystal structure of sulfatase from Pedobacter yulinensis | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | Authors: | O'Malley, A, Schlachter, C.R, Grimes, L.L, Tomashek, J.J, Lee, A.L, Chruszcz, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis .
Molecules, 27, 2021
|
|
8AVQ
 
 | | AO75L in Complex with UDP-Xylose | | Descriptor: | 1,2-ETHANEDIOL, AO75L, BICINE, ... | | Authors: | Laugeri, M.E, Speciale, I, Gimeno, A, Lin, S, Poveda, A, Lowary, T, Van Etten, J.L, Barbero, J.J, De Castro, C, Tonetti, M, Rojas, A.L. | | Deposit date: | 2022-08-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AO75L in Complex with UDP-Xylose
To Be Published
|
|
4JZV
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - second guanosine residue in guanosine binding pocket | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, RNA (5'-R(*(GCP)P*G)-3'), ... | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3MG9
 
 | | Teg 12 Binary Structure Complexed with the Teicoplanin Aglycone | | Descriptor: | FORMIC ACID, GLYCEROL, TEG12, ... | | Authors: | Bick, M.J, Banik, J.J, Darst, S.A, Brady, S.F. | | Deposit date: | 2010-04-05 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structures of the Glycopeptide Sulfotransferase Teg12 in a Complex with the Teicoplanin Aglycone.
Biochemistry, 49, 2010
|
|
4JYZ
 
 | |
1T6W
 
 | | RATIONAL DESIGN OF A CALCIUM-BINDING ADHESION PROTEIN NMR, 20 STRUCTURES | | Descriptor: | CALCIUM ION, hypothetical protein XP_346638 | | Authors: | Yang, W, Wilkins, A.L, Ye, Y, Liu, Z.-R, Urbauer, J.L, Kearney, A, van der Merwe, P.A, Yang, J.J. | | Deposit date: | 2004-05-07 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Design of a calcium-binding protein with desired structure in a cell adhesion molecule.
J.Am.Chem.Soc., 127, 2005
|
|
3ZCO
 
 | | Crystal structure of S. cerevisiae Sir3 C-terminal domain | | Descriptor: | REGULATORY PROTEIN SIR3 | | Authors: | Oppikofer, M, Kueng, S, Keusch, J.J, Hassler, M, Ladurner, A.G, Gut, H, Gasser, S.M. | | Deposit date: | 2012-11-21 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dimerization of Sir3 Via its C-Terminal Winged Helix Domain is Essential for Yeast Heterochromatin Formation.
Embo J., 32, 2013
|
|
4JYK
 
 | | Structure of E. coli Transcriptional Regulator RutR with bound uracil | | Descriptor: | HTH-type transcriptional regulator RutR, SULFATE ION, URACIL | | Authors: | Cooper, D.R, Knapik, A.A, Petkowski, J.J, Porebski, P.J, Osinski, T, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-29 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of E. coli Transcriptional Regulator RutR with bound uracil
To be Published
|
|
4L7D
 
 | | Structure of keap1 kelch domain with (1S,2R)-2-{[(1S)-5-methyl-1-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid | | Descriptor: | (1S,2R)-2-{[(1S)-5-methyl-1-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Jnoff, E, Brookfield, F, Albrecht, C, Barker, J.J, Barker, O, Beaumont, E, Bromidge, S, Brooks, M, Ceska, T, Courade, J.P, Crabbe, T, Duclos, S, Fryatt, T, Jigorel, E, Kwong, J, Sands, Z, Smith, M.A. | | Deposit date: | 2013-06-13 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Binding Mode and Structure-Activity Relationships around Direct Inhibitors of the Nrf2-Keap1 Complex.
Chemmedchem, 9, 2014
|
|
