4ER3
 
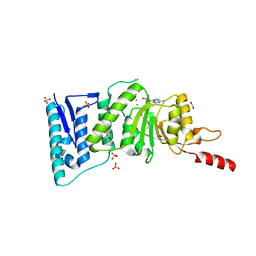 | | Crystal Structure of Human DOT1L in complex with inhibitor EPZ004777 | | Descriptor: | 1,2-ETHANEDIOL, 7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Histone-lysine N-methyltransferase, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Federation, A, Marineau, J, Qi, J, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
1DIB
 
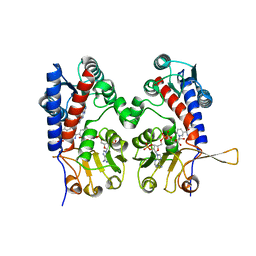 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY345899 | | Descriptor: | 4-(7-AMINO-9-HYDROXY-1-OXO-3,3A,4,5-TETRAHYDRO-2,5,6,8,9B-PENTAAZA-CYCLOPENTA[A]NAPHTHALEN-2-YL)-PHENYLCARBONYL-GLUTAMI C ACID, METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | Deposit date: | 1999-11-29 | | Release date: | 2000-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
1DIG
 
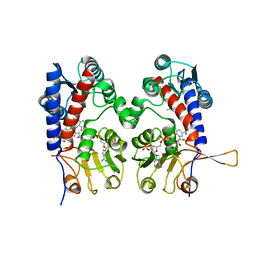 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY374571 | | Descriptor: | ACETATE ION, METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | Deposit date: | 1999-11-29 | | Release date: | 2000-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
1HFD
 
 | | HUMAN COMPLEMENT FACTOR D IN A P21 CRYSTAL FORM | | Descriptor: | COMPLEMENT FACTOR D | | Authors: | Jing, H, Babu, Y.S, Moore, D, Kilpatrick, J.M, Liu, X.-Y, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity.
J.Mol.Biol., 282, 1998
|
|
2P7F
 
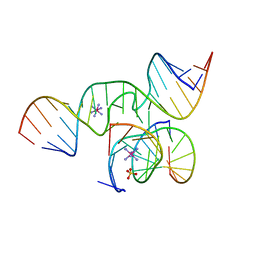 | | The Novel Use of a 2',5'-Phosphodiester Linkage as a Reaction Intermediate at the Active Site of a Small Ribozyme | | Descriptor: | COBALT HEXAMMINE(III), Loop A ribozyme strand, Loop B S-turn strand, ... | | Authors: | Torelli, A.T, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-03-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A comparison of vanadate to a 2'-5' linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization
Rna, 13, 2007
|
|
2P7E
 
 | | Vanadate at the Active Site of a Small Ribozyme Suggests a Role for Water in Transition-State Stabilization | | Descriptor: | 3' substrate strand, octameric fragment, 5' substrate strand, ... | | Authors: | Torelli, A.T, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-03-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A comparison of vanadate to a 2'-5' linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization
Rna, 13, 2007
|
|
1DIA
 
 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY249543 | | Descriptor: | METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [[[2-AMINO-5,6,7,8-TETRAHYDRO-4-HYDROXY-PYRIDO[2,3-D]PYRIMIDIN-6-YL]-ETHYL]-PHENYL]-CARBONYL-GLUTAMIC ACID | | Authors: | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | Deposit date: | 1999-11-29 | | Release date: | 2000-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
2P7D
 
 | | A Minimal, 'Hinged' Hairpin Ribozyme Construct Solved with Mimics of the Product Strands at 2.25 Angstroms Resolution | | Descriptor: | 3' substrate strand, octameric fragment, 5' substrate strand, ... | | Authors: | Torelli, A.T, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2007-03-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A comparison of vanadate to a 2'-5' linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization
Rna, 13, 2007
|
|
6OZ9
 
 | | Ebola virus glycoprotein in complex with EBOV-520 Fab | | Descriptor: | EBOV-520 Fab heavy chain, EBOV-520 Fab light chain, Envelope glycoprotein, ... | | Authors: | Milligan, J.C, Altman, P.X, Hui, S, Hastie, K.M, Gilchuk, P, Crowe, J.E, Saphire, E.O. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.462 Å) | | Cite: | Analysis of a Therapeutic Antibody Cocktail Reveals Determinants for Cooperative and Broad Ebolavirus Neutralization.
Immunity, 52, 2020
|
|
5WHT
 
 | | Crystal structure of 3'SL bound PltB | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Gao, X, Galan, J.E. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Evolution of host adaptation in the Salmonella typhoid toxin.
Nat Microbiol, 2, 2017
|
|
6ZIU
 
 | | bovine ATP synthase stator domain, state 3 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase protein 8, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.02 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2PPI
 
 | | Structure of the BTB (Tramtrack and Bric a brac) domain of human Gigaxonin | | Descriptor: | Gigaxonin | | Authors: | Amos, A, Turnbull, A.P, Tickle, J, Keates, T, Bullock, A, Savitsky, P, Burgess-Brown, N, Debreczeni, J.E, Ugochukwu, E, Umeano, C, Pike, A.C.W, Papagrigoriou, E, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-04-30 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the BTB (Tramtrack and Bric a brac) domain of human Gigaxonin.
To be Published
|
|
7C3M
 
 | | Structure of FERM protein | | Descriptor: | Fermitin family homolog 3,Fermitin family homolog 3,Fermitin family homolog 3 | | Authors: | Bu, W, Loh, Z.Y, Jin, S, Basu, S, Ero, R, Park, J.E, Yan, X, Wang, M, Sze, S.K, Tan, S.M, Gao, Y.G. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of human full-length kindlin-3 homotrimer in an auto-inhibited state.
Plos Biol., 18, 2020
|
|
1COW
 
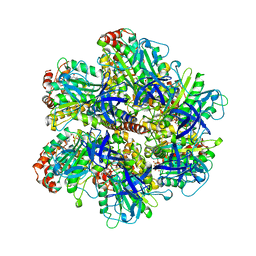 | | BOVINE MITOCHONDRIAL F1-ATPASE COMPLEXED WITH AUROVERTIN B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AUROVERTIN B, BOVINE MITOCHONDRIAL F1-ATPASE, ... | | Authors: | van Raaij, M.J, Abrahams, J.P, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 1996-05-08 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
5WCM
 
 | | Crystal structure of the complex between class B3 beta-lactamase BJP-1 and 4-nitrobenzene-sulfonamide - new refinement | | Descriptor: | 4-nitrobenzenesulfonamide, Blr6230 protein, ZINC ION | | Authors: | Docquier, J.D, Benvenuti, M, Calderone, V, Menciassi, N, Shabalin, I.G, Raczynska, J.E, Wlodawer, A, Jaskolski, M, Minor, W, Mangani, S. | | Deposit date: | 2017-06-30 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structure of the subclass B3 metallo-beta-lactamase BJP-1: rational basis for substrate specificity and interaction with sulfonamides.
Antimicrob. Agents Chemother., 54, 2010
|
|
7CRC
 
 | | Cryo-EM structure of plant NLR RPP1 tetramer in complex with ATR1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Avirulence protein ATR1, ... | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
5WHU
 
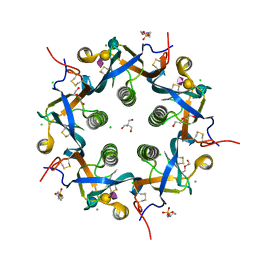 | | Crystal structure of 3'SL bound ArtB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ArtB protein, ... | | Authors: | Gao, X, Galan, J.E. | | Deposit date: | 2017-07-18 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of host adaptation in the Salmonella typhoid toxin.
Nat Microbiol, 2, 2017
|
|
2Q80
 
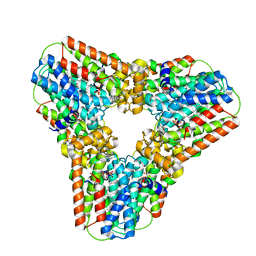 | | Crystal structure of human geranylgeranyl pyrophosphate synthase bound to GGPP | | Descriptor: | GERANYLGERANYL DIPHOSPHATE, Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Kavanagh, K.L, Dunford, J.E, Bunkoczi, G, Smee, C, von Delft, F, Arrowsmith, C, Weigelt, J, Edwards, A, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-08 | | Release date: | 2007-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of human geranylgeranyl pyrophosphate synthase reveals a novel hexameric arrangement and inhibitory product binding
J.Biol.Chem., 281, 2006
|
|
2D2K
 
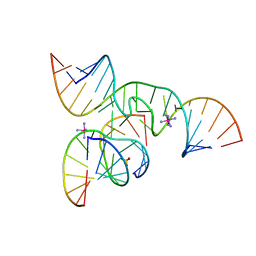 | | Crystal Structure of a minimal, native (U39) all-RNA hairpin ribozyme | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2005-09-11 | | Release date: | 2005-11-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn
Biochemistry, 44, 2005
|
|
2D2L
 
 | | Crystal Structure of a minimal, all-RNA hairpin ribozyme with a propyl linker (C3) at position U39 | | Descriptor: | 5'-R(*CP*GP*GP*UP*GP*AP*GP*AP*AP*GP*GP*G)-3', 5'-R(*GP*GP*CP*AP*GP*AP*GP*AP*AP*AP*CP*AP*CP*AP*CP*GP*A)-3', 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*G)-3', ... | | Authors: | Alam, S, Grum-Tokars, V, Krucinska, J, Kundracik, M.L, Wedekind, J.E. | | Deposit date: | 2005-09-11 | | Release date: | 2005-11-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn
Biochemistry, 44, 2005
|
|
6Z9W
 
 | | Human Class I Major Histocompatibility Complex, A02 allele, presenting LLGWVFAQV | | Descriptor: | Beta-2-microglobulin, LEU-LEU-GLY-TRP-VAL-PHE-ALA-GLN-VAL, MHC class I antigen | | Authors: | Rizkallah, P.J, Man, S, Redman, J.E. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
J Immunol., 207, 2021
|
|
6Z9V
 
 | | Human Class I Major Histocompatibility Complex, A02 allele, presenting IIGWMWIPV | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Rizkallah, P.J, Man, S, Redman, J.E. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
J Immunol., 207, 2021
|
|
6Z9X
 
 | | Human Class I Major Histocompatibility Complex, A02 allele, presenting LLS (t-butyl)Y FGTPT | | Descriptor: | Beta-2-microglobulin, LEU-LEU-SER-TUR-PHE-GLY-THR-PRO-THR, MHC class I antigen, ... | | Authors: | Rizkallah, P.J, Man, S, Redman, J.E. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
J Immunol., 207, 2021
|
|
5W8W
 
 | | Bacillus cereus Zn-dependent metallo-beta-lactamase at pH 7 - new refinement | | Descriptor: | Metallo-beta-lactamase type 2, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Gonzalez, J.M, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Wlodawer, A, Gonzalez, M.M, Vila, A.J. | | Deposit date: | 2017-06-22 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5WEQ
 
 | | The crystal structure of a MR78 mutant | | Descriptor: | MR78 mutant Fab heavy chain, MR78 mutant light chain | | Authors: | Dong, J, Williamson, L.E, Crowe, J.E. | | Deposit date: | 2017-07-10 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of Non-local Interactions between CDR Loops in Binding Affinity of MR78 Antibody to Marburg Virus Glycoprotein.
Structure, 25, 2017
|
|
