5LRX
 
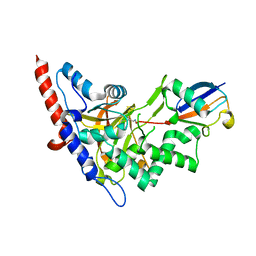 | | Structure of A20 OTU domain bound to ubiquitin | | Descriptor: | Polyubiquitin-B, Tumor necrosis factor alpha-induced protein 3 | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
5NN2
 
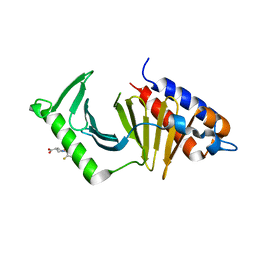 | | The structure of the polo-box domain (PBD) of Plk1 in complex with Z228588490 | | Descriptor: | (3~{R},5~{R})-1-[2,4-bis(fluoranyl)phenyl]-5-oxidanyl-pyrrolidine-3-carboxylic acid, Serine/threonine-protein kinase PLK1 | | Authors: | Kunciw, D.L, Rossmann, M, Stokes, J.E, De Fusco, C, Spring, D.R, Hyvonen, M. | | Deposit date: | 2017-04-07 | | Release date: | 2018-02-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A fragment-based approach to developing inhibitors of the cryptic pocket of the Polo-Like Kinase 1 Polo-Box Domain.
To Be Published
|
|
5NHH
 
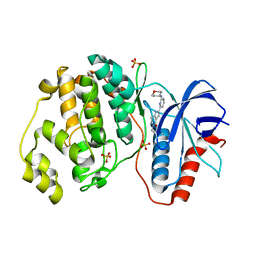 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 5-(2-methoxyethyl)-2-[2-(oxan-4-ylamino)pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHO
 
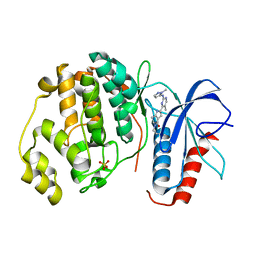 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | (6~{S})-5-(2-methoxyethyl)-6-methyl-2-[5-methyl-2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
2Y6S
 
 | | Structure of an Ebolavirus-protective antibody in complex with its mucin-domain linear epitope | | Descriptor: | ENVELOPE GLYCOPROTEIN, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Olal, D.O, Kuehne, A, Lee, J.E, Bale, S, Dye, J.M, Saphire, E.O. | | Deposit date: | 2011-01-25 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an Ebola Virus-Protective Antibody in Complex with its Mucin-Domain Linear Epitope.
J.Virol., 86, 2012
|
|
2YCQ
 
 | | Crystal structure of checkpoint kinase 2 in complex with inhibitor PV1115 | | Descriptor: | N-{4-[(1E)-N-1H-IMIDAZOL-2-YLETHANEHYDRAZONOYL]PHENYL}-7-NITRO-1H-INDOLE-2-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C.R, Pommier, Y, Shoemaker, R.H, Zhang, G, Waugh, D.S. | | Deposit date: | 2011-03-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
5NHV
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 7-[2-(oxan-4-ylamino)pyrimidin-4-yl]-3,4-dihydro-2~{H}-pyrrolo[1,2-a]pyrazin-1-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
1K0E
 
 | | THE CRYSTAL STRUCTURE OF AMINODEOXYCHORISMATE SYNTHASE FROM FORMATE GROWN CRYSTALS | | Descriptor: | FORMIC ACID, TRYPTOPHAN, p-aminobenzoate synthase component I | | Authors: | Parsons, J.F, Jensen, P.Y, Pachikara, A.S, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli aminodeoxychorismate synthase: architectural conservation and diversity in chorismate-utilizing enzymes.
Biochemistry, 41, 2002
|
|
1JYO
 
 | |
1ORY
 
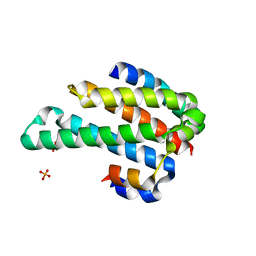 | | FLAGELLAR EXPORT CHAPERONE IN COMPLEX WITH ITS COGNATE BINDING PARTNER | | Descriptor: | Flagellin, PHOSPHATE ION, flagellar protein FliS | | Authors: | Evdokimov, A.G, Phan, J, Tropea, J.E, Routzahn, K.M, Peters III, H.K, Pokross, M, Waugh, D.S. | | Deposit date: | 2003-03-17 | | Release date: | 2003-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion
Nat.Struct.Biol., 10, 2003
|
|
3G1R
 
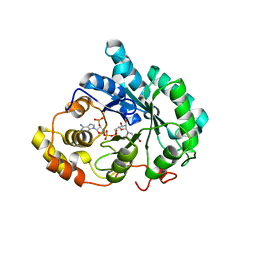 | | Crystal structure of human liver 5beta-reductase (AKR1D1) in complex with NADP and Finasteride. Resolution 1.70 A | | Descriptor: | (4aR,4bS,6aS,7S,9aS,9bS,11aR)-N-tert-butyl-4a,6a-dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxamide, 3-oxo-5-beta-steroid 4-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Di Costanzo, L, Drury, J.E, Penning, T.M, Christianson, D.W. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Inhibition of human steroid 5beta-reductase (AKR1D1) by finasteride and structure of the enzyme-inhibitor complex.
J.Biol.Chem., 284, 2009
|
|
1K0G
 
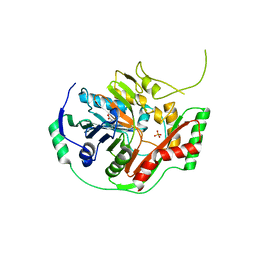 | | THE CRYSTAL STRUCTURE OF AMINODEOXYCHORISMATE SYNTHASE FROM PHOSPHATE GROWN CRYSTALS | | Descriptor: | PHOSPHATE ION, TRYPTOPHAN, p-aminobenzoate synthase component I | | Authors: | Parsons, J.F, Jensen, P.Y, Pachikara, A.S, Howard, A.J, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Escherichia coli aminodeoxychorismate synthase: architectural conservation and diversity in chorismate-utilizing enzymes.
Biochemistry, 41, 2002
|
|
3G4Y
 
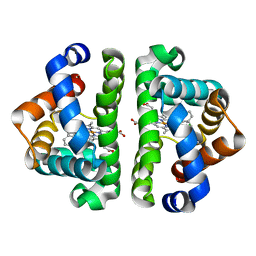 | | Ligand migration and cavities within scapharca dimeric hemoglobin: wild type with co bound to heme and chloromethyl benzene bound to the XE4 cavity | | Descriptor: | (chloromethyl)benzene, CARBON MONOXIDE, GLOBIN-1, ... | | Authors: | Knapp, J.E, Pahl, R, Cohen, J, Nichols, J.C, Schulten, K, Gibson, Q.H, Srajer, V, Royer Jr, W.E. | | Deposit date: | 2009-02-04 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand migration and cavities within Scapharca Dimeric HbI: studies by time-resolved crystallo-graphy, Xe binding, and computational analysis.
Structure, 17, 2009
|
|
3G53
 
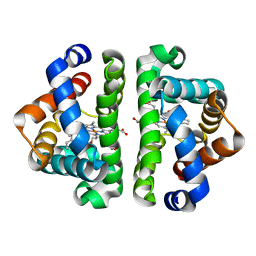 | | Ligand migration and cavities within scapharca dimeric hemoglobin: wild type with co bound to heme and chloropropyl benzene bound to the XE4 cavity | | Descriptor: | (3-chloropropyl)benzene, CARBON MONOXIDE, Globin-1, ... | | Authors: | Knapp, J.E, Pahl, R, Cohen, J, Nichols, J.C, Schulten, K, Gibson, Q.H, Srajer, V, Royer Jr, W.E. | | Deposit date: | 2009-02-04 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ligand migration and cavities within Scapharca Dimeric HbI: studies by time-resolved crystallo-graphy, Xe binding, and computational analysis.
Structure, 17, 2009
|
|
5NGU
 
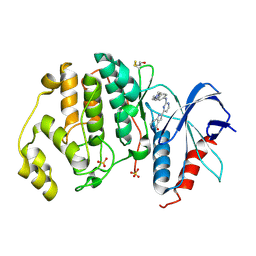 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 2-[2-[[4-(4-methylpiperazin-1-yl)phenyl]amino]pyrimidin-4-yl]-1,5,6,7-tetrahydropyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
7RVB
 
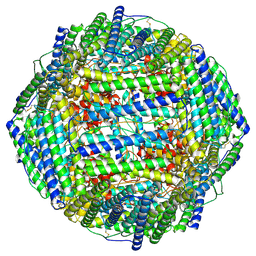 | | High resolution map of molecular chaperone Artemin | | Descriptor: | Ferritin | | Authors: | Parvate, A.D, Powell, S.M, Brookreason, J.T, Novikova, I.V, Evans, J.E. | | Deposit date: | 2021-08-18 | | Release date: | 2022-12-14 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Cryo-EM structure of the diapause chaperone artemin.
Front Mol Biosci, 9, 2022
|
|
1P9J
 
 | | Solution structure and dynamics of the EGF/TGF-alpha chimera T1E | | Descriptor: | chimera of Epidermal growth factor(EGF) and Transforming growth factor alpha (TGF-alpha) | | Authors: | Wingens, M, Walma, T, Van Ingen, H, Stortelers, C, Van Leeuwen, J.E, Van Zoelen, E.J, Vuister, G.W. | | Deposit date: | 2003-05-12 | | Release date: | 2003-10-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of an Epidermal Growth Factor/Transforming Growth Factor-alpha Chimera with Unique ErbB Binding Specificity.
J.Biol.Chem., 278, 2003
|
|
1JWN
 
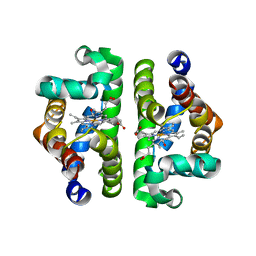 | | Crystal Structure of Scapharca inaequivalvis HbI, I114F Mutant Ligated to Carbon Monoxide. | | Descriptor: | CARBON MONOXIDE, Globin I - Ark Shell, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Gibson, Q.H, Cushing, L, Royer Jr, W.E. | | Deposit date: | 2001-09-04 | | Release date: | 2001-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Restricting the ligand-linked heme movement in Scapharca dimeric hemoglobin reveals tight coupling between distal and proximal contributions to cooperativity.
Biochemistry, 40, 2001
|
|
2NDM
 
 | | NMR solution structure of PawS Derived Peptide 21 (PDP-21) | | Descriptor: | PawS derived peptide 21 | | Authors: | Franke, B, Jayasena, A.S, Fisher, M.F, Swedberg, J.E, Taylor, N.L, Mylne, J.S, Rosengren, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Diverse cyclic seed peptides in the Mexican zinnia (Zinnia haageana).
Biopolymers, 106, 2016
|
|
3I2S
 
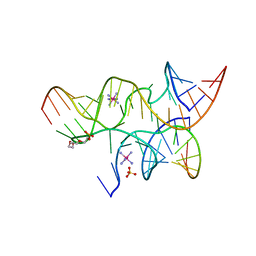 | | Crystal structure of the hairpin ribozyme with a 2'OMe substrate and N1-deazaadenosine at position A10 | | Descriptor: | 5'-R(*UP*CP*CP*CP*(A2M)P*GP*UP*CP*CP*AP*CP*CP*GP*U)-3', 5'-R(*UP*CP*GP*UP*GP*GP*UP*AP*CP*AP*UP*UP*AP*CP*CP*UP*GP*CP*C)-3', COBALT HEXAMMINE(III), ... | | Authors: | Wedekind, J.E, Spitale, R.C, Krucinska, J. | | Deposit date: | 2009-06-29 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Single-atom imino substitutions at A9 and A10 reveal distinct effects on the fold and function of the hairpin ribozyme catalytic core.
Biochemistry, 48, 2009
|
|
2WO1
 
 | | Crystal Structure of the EphA4 Ligand Binding Domain | | Descriptor: | EPHRIN TYPE-A RECEPTOR, N-PROPANOL | | Authors: | Bowden, T.A, Aricescu, A.R, Nettleship, J.E, Siebold, C, Rahman-Huq, N, Owens, R.J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2009-07-21 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Plasticity of Eph-Receptor A4 Facilitates Cross-Class Ephrin Signalling
Structure, 17, 2009
|
|
5N0I
 
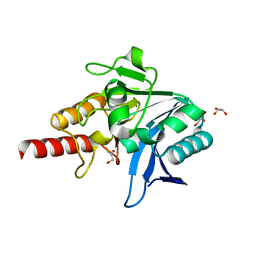 | | Crystal structure of NDM-1 in complex with beta-mercaptoethanol - new refinement | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, King, D.T, Strynadka, N.C.J. | | Deposit date: | 2017-02-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
4D4L
 
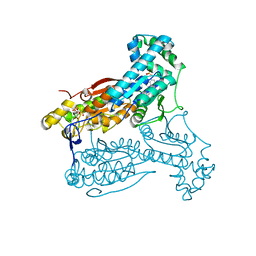 | | human PFKFB3 in complex with a pyrrolopyrimidone compound | | Descriptor: | 5-(4-chlorophenyl)-7-phenyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Stgallay, S.A, Bennett, N, Critchlow, S, Davies, G, Debreczeni, J.E, Evans, N, Holdgate, G, Jones, N.P, Leach, L, Maman, S, McLoughlin, S, Preston, M, Rigoreau, L, Thomas, A, Walker, G, Walsch, J, Ward, R.A, Wheatley, E, Winter, J. | | Deposit date: | 2014-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Identifying a Novel Series of Pfkfb3 Inhibitors as a Metabolic Approach to Treating Cancer from Hts, Biophysical and Biochemical Methods
To be Published
|
|
2X7O
 
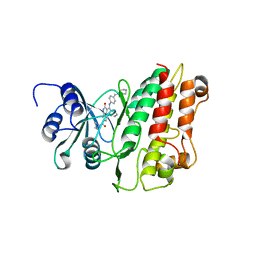 | | Crystal structure of TGFbRI complexed with an indolinone inhibitor | | Descriptor: | (3Z)-N-ETHYL-N-METHYL-2-OXO-3-(PHENYL{[4-(PIPERIDIN-1-YLMETHYL)PHENYL]AMINO}METHYLIDENE)-2,3-DIHYDRO-1H-INDOLE-6-CARBOXAMIDE, TGF-BETA RECEPTOR TYPE I | | Authors: | Roth, G.J, Heckel, A, Brandl, T, Grauert, M, Hoerer, S, Kley, J.T, Schnapp, G, Baum, P, Mennerich, D, Schnapp, A, Park, J.E. | | Deposit date: | 2010-03-03 | | Release date: | 2010-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Design, Synthesis and Evaluation of Indolinones as Inhibitors of the Transforming Growth Factor Beta Receptor I (Tgfbri)
J.Med.Chem., 53, 2010
|
|
2NUG
 
 | | Crystal structure of RNase III from Aquifex aeolicus complexed with ds-RNA at 1.7-Angstrom Resolution | | Descriptor: | 5'-R(P*AP*AP*GP*GP*UP*CP*AP*UP*UP*CP*G)-3', 5'-R(P*AP*GP*UP*GP*GP*CP*CP*UP*UP*GP*C)-3', MAGNESIUM ION, ... | | Authors: | Gan, J.H, Shaw, G, Tropea, J.E, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2006-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A stepwise model for double-stranded RNA processing by ribonuclease III.
Mol.Microbiol., 67, 2007
|
|
