6VXR
 
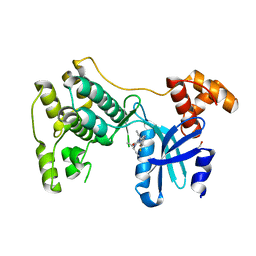 | | Structure of Maternal embryonic leucine zipper kinase bound to LDSM276 | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-5,8-di(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, 1,2-ETHANEDIOL, Maternal embryonic leucine zipper kinase | | Authors: | Counago, R.M, Takarada, J.E, dos Reis, C.V, Gama, F.H, Azevedo, H, Mascarello, A, Guimaraes, C.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-24 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Maternal embryonic leucine zipper kinase bound to LDSM276
To Be Published
|
|
2IZS
 
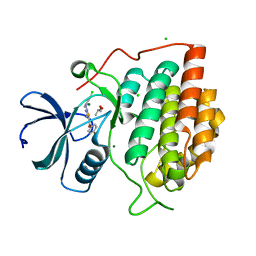 | | Structure of casein kinase gamma 3 in complex with inhibitor | | Descriptor: | CASEIN KINASE I ISOFORM GAMMA-3, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bunkoczi, G, Salah, E, Rellos, P, Das, S, Fedorov, O, Savitsky, P, Debreczeni, J.E, Gileadi, O, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, von Delft, F, Knapp, S. | | Deposit date: | 2006-07-26 | | Release date: | 2006-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibitor Binding by Casein Kinases
To be Published
|
|
2IFY
 
 | |
2IJH
 
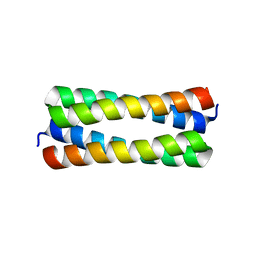 | | Crystal structure analysis of ColE1 ROM mutant F14W | | Descriptor: | Regulatory protein rop | | Authors: | Ladner, J.E. | | Deposit date: | 2006-09-29 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New crystal structures of ColE1 Rom and variants resulting from mutation of a surface exposed residue: Implications for RNA-recognition.
Proteins, 72, 2008
|
|
6XH0
 
 | |
2IME
 
 | | 2-Hydroxychromene-2-carboxylate Isomerase: a Kappa Class Glutathione-S-Transferase from Pseudomonas putida | | Descriptor: | (2S)-2-HYDROXY-2H-CHROMENE-2-CARBOXYLIC ACID, (3E)-4-(2-HYDROXYPHENYL)-2-OXOBUT-3-ENOIC ACID, 2-hydroxychromene-2-carboxylate isomerase, ... | | Authors: | Thompson, L.C, Ladner, J.E, Codreanu, S.G, Harp, J, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2006-10-04 | | Release date: | 2007-06-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Hydroxychromene-2-carboxylic Acid Isomerase: A Kappa Class Glutathione Transferase from Pseudomonas putida
Biochemistry, 46, 2007
|
|
2J2F
 
 | | The T199D Mutant of Stearoyl Acyl Carrier Protein Desaturase from Ricinus Communis (Castor Bean) | | Descriptor: | ACYL-[ACYL-CARRIER-PROTEIN] DESATURASE, FE (III) ION | | Authors: | Guy, J.E, Abreu, I.A, Moche, M, Lindqvist, Y, Whittle, E, Shanklin, J. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Single Mutation in the Castor {Delta}9-18:0- Desaturase Changes Reaction Partitioning from Desaturation to Oxidase Chemistry.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
6XMJ
 
 | | Human 20S proteasome bound to an engineered 11S (PA26) activator | | Descriptor: | Proteasome activator protein PA26, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | de la Pena, A.H, Opoku-Nsiah, K.A, Williams, S.K, Chopra, N, Sali, A, Gestwicki, J.E, Lander, G.C. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Y Phi motif defines the structure-activity relationships of human 20S proteasome activators.
Nat Commun, 13, 2022
|
|
6VYG
 
 | | Cryo-EM structure of Plasmodium vivax hexokinase (Closed state) | | Descriptor: | Phosphotransferase | | Authors: | Srivastava, S.S, Darling, J.E, Suryadi, J, Morris, J.C, Drew, M.E, Subramaniam, S. | | Deposit date: | 2020-02-26 | | Release date: | 2020-05-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Plasmodium vivax and human hexokinases share similar active sites but display distinct quaternary architectures
Iucrj, 7, 2020
|
|
8VPV
 
 | |
5FIK
 
 | | Bovine mitochondrial ATP synthase state 3a | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
5FIJ
 
 | | Bovine mitochondrial ATP synthase state 2c | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
2I0H
 
 | | The structure of p38alpha in complex with an arylpyridazinone | | Descriptor: | 2-(3-{(2-CHLORO-4-FLUOROPHENYL)[1-(2-CHLOROPHENYL)-6-OXO-1,6-DIHYDROPYRIDAZIN-3-YL]AMINO}PROPYL)-1H-ISOINDOLE-1,3(2H)-DIONE, GLYCEROL, Mitogen-activated protein kinase 14 | | Authors: | Natarajan, S.R, Heller, S.T, Nam, K, Singh, S.B, Scapin, G, Patel, S, Thompson, J.E, Fitzgerald, C.E, O'Keefe, S.J. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | p38 MAP Kinase Inhibitors Part 6: 2-Arylpyridazin-3-ones as templates for inhibitor design.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
6VUI
 
 | | Metabolite-bound PreQ1 riboswitch with Mn2+ | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, MANGANESE (II) ION, PREQ1 RIBOSWITCH | | Authors: | Jenkins, J.L, Wedekind, J.E. | | Deposit date: | 2020-02-15 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.681 Å) | | Cite: | Analysis of a preQ1-I riboswitch in effector-free and bound states reveals a metabolite-programmed nucleobase-stacking spine that controls gene regulation.
Nucleic Acids Res., 48, 2020
|
|
2I5S
 
 | | Crystal structure of onconase with bound nucleic acid | | Descriptor: | 5'-D(*A*(DU)P*GP*A)-3', P-30 protein | | Authors: | Bae, E, Lee, J.E, Raines, R.T, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for catalysis by onconase.
J.Mol.Biol., 375, 2008
|
|
6VQ2
 
 | | HLA-B*27:05 presenting an HIV-1 14mer peptide | | Descriptor: | 14-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQE
 
 | | HLA-B*27:05 presenting an HIV-1 13mer peptide | | Descriptor: | 13-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
3UHS
 
 | | HBI (L36M) deoxy | | Descriptor: | Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ren, Z, Srajer, V, Knapp, J.E, Royer Jr, W.E. | | Deposit date: | 2011-11-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5FOY
 
 | | De novo structure of the binary mosquito larvicide BinAB at pH 7 | | Descriptor: | 41.9 KDA INSECTICIDAL TOXIN, LARVICIDAL TOXIN 51 KDA PROTEIN | | Authors: | Colletier, J.P, Sawaya, M.R, Gingery, M, Rodriguez, J.A, Cascio, D, Brewster, A.S, Michels-Clark, T, Boutet, S, Williams, G.J, Messerschmidt, M, DePonte, D.P, Sierra, R.G, Laksmono, H, Koglin, J.E, Hunter, M.S, W Park, H, Uervirojnangkoorn, M, Bideshi, D.L, Brunger, A.T, Federici, B.A, Sauter, N.K, Eisenberg, D.S. | | Deposit date: | 2015-11-26 | | Release date: | 2016-10-05 | | Last modified: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | De Novo Phasing with X-Ray Laser Reveals Mosquito Larvicide Binab Structure.
Nature, 539, 2016
|
|
6VQZ
 
 | | HLA-B*27:05 presenting an HIV-1 6mer peptide | | Descriptor: | 6-mer peptide, ARGININE, Beta-2-microglobulin, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VPZ
 
 | | HLA-B*27:05 presenting an HIV-1 11mer peptide | | Descriptor: | 11-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-04 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQD
 
 | | HLA-B*27:05 presenting an HIV-1 8mer peptide | | Descriptor: | 8-mer peptide, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
6VQY
 
 | | HLA-B*27:05 presenting an HIV-1 7mer peptide | | Descriptor: | 7-mer peptide, ARGININE, Beta-2-microglobulin, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
3UFD
 
 | | C.Esp1396I bound to its highest affinity operator site OM | | Descriptor: | CHLORIDE ION, DNA (5'-D(*AP*TP*GP*TP*AP*GP*AP*CP*TP*AP*TP*AP*GP*TP*CP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*GP*AP*CP*TP*AP*TP*AP*GP*TP*CP*TP*AP*CP*A)-3'), ... | | Authors: | Ball, N.J, McGeehan, J.E, Streeter, S.D, Thresh, S.-J, Kneale, G.G. | | Deposit date: | 2011-11-01 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of differential DNA sequence recognition by restriction-modification controller proteins.
Nucleic Acids Res., 40, 2012
|
|
6W7L
 
 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ632p | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-phenylimidazo[1,2-a]pyrazin-3-yl)amino]benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.856 Å) | | Cite: | Small molecule microarray identifies inhibitors of tyrosyl-DNA phosphodiesterase 1 that simultaneously access the catalytic pocket and two substrate binding sites
Chemical Science, 12, 2021
|
|
