4KAG
 
 | | Crystal structure analysis of a single amino acid deletion mutation in EGFP | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Green fluorescent protein, ... | | Authors: | Arpino, J.A.J, Rizkallah, P.J. | | Deposit date: | 2013-04-22 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural and dynamic changes associated with beneficial engineered single-amino-acid deletion mutations in enhanced green fluorescent protein.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5L93
 
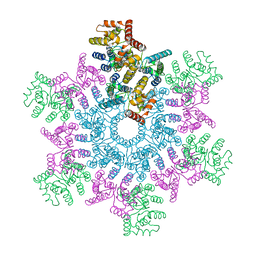 | | An atomic model of HIV-1 CA-SP1 reveals structures regulating assembly and maturation | | Descriptor: | Capsid protein p24 | | Authors: | Schur, F.K.M, Obr, M, Hagen, W.J.H, Wan, W, Arjen, J.J, Kirkpatrick, J.M, Sachse, C, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation.
Science, 353, 2016
|
|
4KA9
 
 | |
8SKB
 
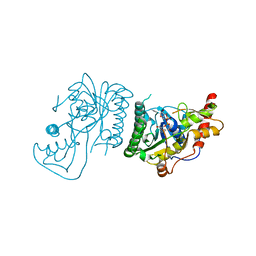 | | Crystal Structure of GDP-mannose 3,5 epimerase de Myrciaria dubia in complex with NAD | | Descriptor: | GDP-mannose 3,5 epimerase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-04-19 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
8SCC
 
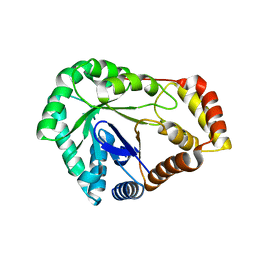 | | Crystal Structure of L-galactose 1-dehydrogenase de Myrciaria dubia | | Descriptor: | L-galactose dehydrogenase | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-04-05 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
8SG0
 
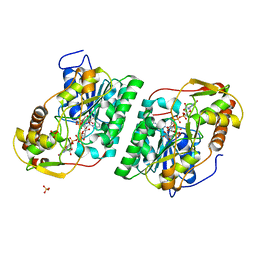 | | Crystal Structure of GDP-manose 3,5 epimerase de Myrciaria dubia in complex with substrate, product and NAD | | Descriptor: | GDP-mannose 3,5-epimerase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, GUANOSINE-5'-DIPHOSPHATE-BETA-L-GALACTOSE, ... | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
5KZD
 
 | | N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus with bound sialic acid alditol | | Descriptor: | (2~{S},4~{S},5~{R},6~{R},7~{S},8~{R})-5-acetamido-2,4,6,7,8,9-hexakis(oxidanyl)nonanoic acid, N-acetylneuraminate lyase | | Authors: | North, R.A, Watson, A.J.A, Pearce, F.G, Muscroft-Taylor, A.C, Friemann, R, Fairbanks, A.J, Dobson, R.C.J. | | Deposit date: | 2016-07-25 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Structure and inhibition of N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus.
FEBS Lett., 590, 2016
|
|
1E4P
 
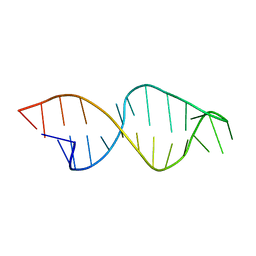 | |
7XU2
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU0
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-211 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XTZ
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU1
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU4
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU5
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU3
 
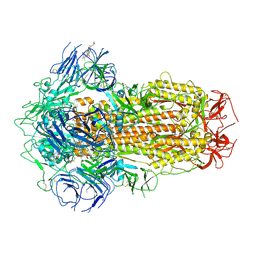 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU6
 
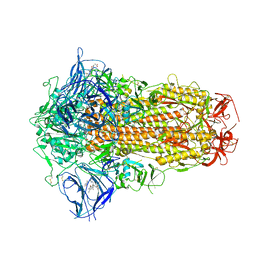 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), incubated in Low pH after 40-Day Storage in PBS, Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
6VPS
 
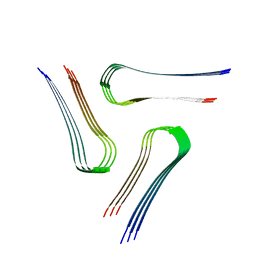 | | Cryo-EM structure of the amyloid core of Drosophila Orb2 isolated from head | | Descriptor: | Translational regulator orb2 | | Authors: | Hervas, R, Rau, M.J, Park, Y, Zhang, W, Murzin, A.G, Fitzpatrick, J.A.J, Scheres, S.H.W, Si, K. | | Deposit date: | 2020-02-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila
Science, 367, 2020
|
|
6MRQ
 
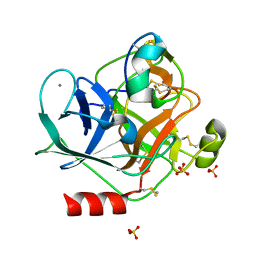 | | Structure of ToPI1 inhibitor from Tityus obscurus scorpion venom in complex with trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Fernandes, J.C, Mourao, C.B.F, Schwartz, E.F, Barbosa, J.A.R.G. | | Deposit date: | 2018-10-15 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Head-to-Tail Cyclization after Interaction with Trypsin: A Scorpion Venom Peptide that Resembles Plant Cyclotides.
J.Med.Chem., 63, 2020
|
|
4BLF
 
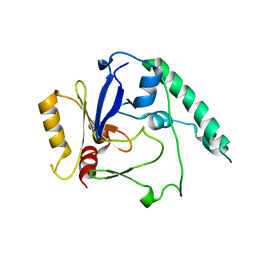 | | Variable internal flexibility characterizes the helical capsid formed by Agrobacterium VirE2 protein on single-stranded DNA. | | Descriptor: | SINGLE-STRAND DNA-BINDING PROTEIN | | Authors: | Bharat, T.A.M, Zbaida, D, Eisenstein, M, Frankenstein, Z, Mehlman, T, Weiner, L, Sorzano, C.O.S, Barak, Y, Albeck, S, Briggs, J.A.G, Wolf, S.G, Elbaum, M. | | Deposit date: | 2013-05-02 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Variable Internal Flexibility Characterizes the Helical Capsid Formed by Agrobacterium Vire2 Protein on Single-Stranded DNA.
Structure, 21, 2013
|
|
4BBO
 
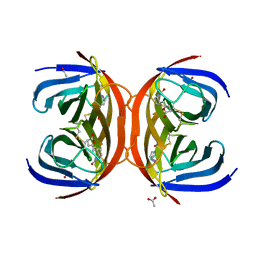 | | Crystal structure of core-bradavidin | | Descriptor: | ACETATE ION, BIOTIN, BLR5658 PROTEIN, ... | | Authors: | Airenne, T.T, Johnson, M.S, Maatta, J.A.E, Hytonen, V.H, Kulomaa, M.S. | | Deposit date: | 2012-09-27 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Core-Bradavidin
To be Published
|
|
5A1U
 
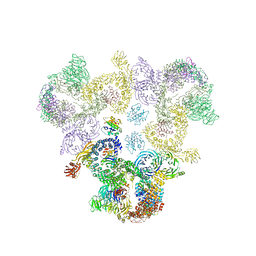 | | The structure of the COPI coat triad | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
5A9E
 
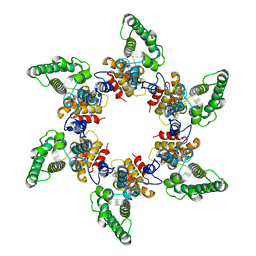 | | Cryo-electron tomography and subtomogram averaging of Rous-Sarcoma- Virus deltaMBD virus-like particles | | Descriptor: | DELTAMBD GAG PROTEIN | | Authors: | Schur, F.K.M, Dick, R.A, Hagen, W.J.H, Vogt, V.M, Briggs, J.A.G. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | The Structure of Immature-Like Rous Sarcoma Virus Gag Particles Reveals a Structural Role for the P10 Domain in Assembly.
J.Virol., 89, 2015
|
|
5A1Y
 
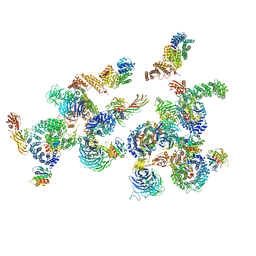 | | The structure of the COPI coat linkage IV | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
5A1V
 
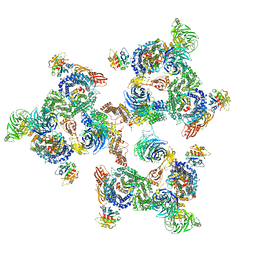 | | The structure of the COPI coat linkage I | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
5A1W
 
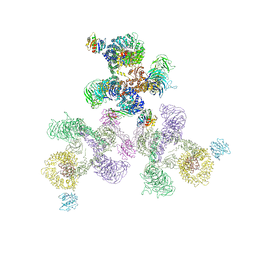 | | The structure of the COPI coat linkage II | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
