3UXW
 
 | | Crystal Structures of an A-T-hook/DNA complex | | Descriptor: | A-T hook peptide, dodecamer DNA | | Authors: | Fonfria-Subiros, E, Acosta-Reyes, F.J, Saperas, N, Pous, J, Subirana, J.A, Campos, J.L. | | Deposit date: | 2011-12-05 | | Release date: | 2012-05-23 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of a complex of DNA with one AT-hook of HMGA1.
Plos One, 7, 2012
|
|
1RXV
 
 | | Crystal Structure of A. Fulgidus FEN-1 bound to DNA | | Descriptor: | 5'-d(*T*pA*pG*pC*pA*pT*pC*pG*pG), Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
7JT4
 
 | | Crystal Structure of BPTF bromodomain labelled with 5-fluoro-tryptophan | | Descriptor: | Nucleosome-remodeling factor subunit BPTF | | Authors: | Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2020-08-17 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Selectivity, ligand deconstruction, and cellular activity analysis of a BPTF bromodomain inhibitor
Org.Biomol.Chem., 17, 2019
|
|
3G1H
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 5,6-dihydrouridine 5'-monophosphate | | Descriptor: | 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
1S23
 
 | | Crystal Structure Analysis of the B-DNA Decamer CGCAATTGCG | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*TP*TP*GP*CP*G)-3', COBALT (II) ION | | Authors: | Valls, N, Wright, G, Steiner, R.A, Murshudov, G.N, Subirana, J.A. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA variability in five crystal structures of d(CGCAATTGCG).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5A4C
 
 | | FGFR1 ligand complex | | Descriptor: | 1,2-ETHANEDIOL, 1-tert-butyl-3-[2-[3-(diethylamino)propylamino]-6-(3,5-dimethoxyphenyl)pyrido[2,3-d]pyrimidin-7-yl]urea, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Klein, T, Vajpai, N, Phillips, J.J, Davies, G, Holdgate, G.A, Phillips, C, Tucker, J.A, Norman, R.A, Scott, A.S, Higazi, D.R, Lowe, D, Thompson, G.S, Breeze, A.L. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Dynamic Insights Into the Energetics of Activation Loop Rearrangement in Fgfr1 Kinase.
Nat.Commun., 6, 2015
|
|
7JRS
 
 | |
3V5C
 
 | | Crystal structure of the mutant E234A of Galacturonate Dehydratase from GEOBACILLUS SP. complexed with Mg | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Groninger-Poe, F, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-12-16 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of the mutant E234A of Galacturonate Dehydratase from GEOBACILLUS SP. complexed with Mg
To be Published
|
|
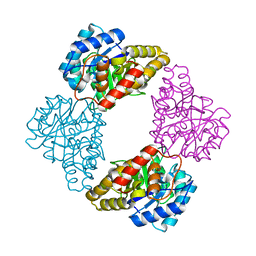
1S5T
 
 | |
3ZE5
 
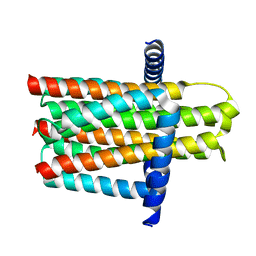 | | Crystal structure of the integral membrane diacylglycerol kinase - delta4 | | Descriptor: | DIACYLGLYCEROL KINASE | | Authors: | Li, D, Vogeley, L, Pye, V.E, Lyons, J.A, Aragao, D, Caffrey, M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Crystal Structure of the Integral Membrane Diacylglycerol Kinase.
Nature, 497, 2013
|
|
3ZE3
 
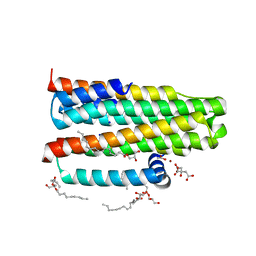 | | Crystal structure of the integral membrane diacylglycerol kinase - delta7 | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ACETATE ION, ... | | Authors: | Li, D, Pye, V.E, Lyons, J.A, Vogeley, L, Aragao, D, Caffrey, M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the Integral Membrane Diacylglycerol Kinase.
Nature, 497, 2013
|
|
3ZE4
 
 | | Crystal structure of the integral membrane diacylglycerol kinase - wild-type | | Descriptor: | DIACYLGLYCEROL KINASE | | Authors: | Li, D, Lyons, J.A, Pye, V.E, Vogeley, L, Aragao, D, Caffrey, M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.702 Å) | | Cite: | Crystal Structure of the Integral Membrane Diacylglycerol Kinase.
Nature, 497, 2013
|
|
3DZ6
 
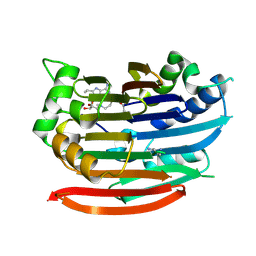 | | Human AdoMetDC with 5'-[(4-aminooxybutyl)methylamino]-5'deoxy-8-ethyladenosine | | Descriptor: | 1,4-DIAMINOBUTANE, 5'-{[4-(aminooxy)butyl](methyl)amino}-5'-deoxy-8-ethenyladenosine, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Bale, S, McCloskey, D.E, Pegg, A.E, Secrist III, J.A, Guida, W.C, Ealick, S.E. | | Deposit date: | 2008-07-29 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | New Insights into the Design of Inhibitors of Human S-Adenosylmethionine Decarboxylase: Studies of Adenine C8 Substitution in Structural Analogues of S-Adenosylmethionine
J.Med.Chem., 52, 2009
|
|
7JO9
 
 | | 1:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
1SH2
 
 | | Crystal Structure of Norwalk Virus Polymerase (Metal-free, Centered Orthorhombic) | | Descriptor: | RNA Polymerase | | Authors: | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
1SFF
 
 | | Structure of gamma-aminobutyrate aminotransferase complex with aminooxyacetate | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-ACETYLYAMINO-PYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Carter, R.J, Zhou, X, Langston, J.A, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-02-19 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of unbound and aminooxyacetate-bound Escherichia coli gamma-aminobutyrate aminotransferase.
Biochemistry, 43, 2004
|
|
3ZC3
 
 | | FERREDOXIN-NADP REDUCTASE (MUTATION S80A) COMPLEXED WITH NADP BY COCRYSTALLIZATION | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Martinez-Julvez, M, Hurtado-Guerrero, R, Herguedas, B, Sanchez-Azqueta, A, Hervas, M, Navarro, J.A, Medina, M. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Hydrogen Bond Network in the Active Site of Anabaena Ferredoxin-Nadp(+) Reductase Modulates its Catalytic Efficiency.
Biochim.Biophys.Acta, 1837, 2013
|
|
3ZDJ
 
 | |
5AIG
 
 | | Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. Tomsk-sample- Valpromide complex | | Descriptor: | 2-PROPYLPENTANAMIDE, DIMETHYL SULFOXIDE, LIMONENE-1,2-EPOXIDE HYDROLASE | | Authors: | Ferrandi, E, Sayer, C, Isupov, M.N, Annovazzi, C, Marchesi, C, Iacobone, G, Peng, X, Bonch-Osmolovskaya, E, Wohlgemuth, R, Littlechild, J.A, Montia, D. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Discovery and Characterization of Thermophilic Limonene-1,2-Epoxide Hydrolases from Hot Spring Metagenomic Libraries
FEBS J., 282, 2015
|
|
3ZIV
 
 | |
1RXW
 
 | | Crystal structure of A. fulgidus FEN-1 bound to DNA | | Descriptor: | 5'-d(*C*pG*pA*pT*pG*pC*pT*pA)-3', 5'-d(*T*pA*pG*pC*pA*pT*pC*pG*pG)-3', Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
3ZJV
 
 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3213 in the editing conformation | | Descriptor: | LEUCINE--TRNA LIGASE, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
1RXZ
 
 | | C-terminal region of A. fulgidus FEN-1 complexed with A. fulgidus PCNA | | Descriptor: | DNA polymerase sliding clamp, Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
3ZQR
 
 | | NMePheB25 insulin analogue crystal structure | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Antolikova, E, Zakova, L, Turkenburg, J.P, Watson, C.J, Hanclova, I, Sanda, M, Cooper, A, Kraus, T, Brzozowski, A.M, Jiracek, J.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Non-Equivalent Role of Inter- and Intramolecular Hydrogen Bonds in the Insulin Dimer Interface.
J.Biol.Chem., 286, 2011
|
|
7K2V
 
 | |
