3UOT
 
 | | Crystal Structure of MDC1 FHA Domain in Complex with a Phosphorylated Peptide from the MDC1 N-terminus | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Clapperton, J.A, Lloyd, J, Haire, L.F, Li, J, Smerdon, S.J. | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator.
Nucleic Acids Res., 40, 2012
|
|
4ZNJ
 
 | | Thermus Phage P74-26 Large Terminase ATPase domain mutant R139A (I 2 3 space group) | | Descriptor: | Phage terminase large subunit, SULFATE ION | | Authors: | Hilbert, B.J, Hayes, J.A, Stone, N.P, Duffy, C.M, Kelch, B.A. | | Deposit date: | 2015-05-04 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Structure and mechanism of the ATPase that powers viral genome packaging.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZNU
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 2-Methyl-substituted OBHS derivative | | Descriptor: | 2-methylphenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4ZL4
 
 | | Plasmepsin V from Plasmodium vivax bound to a transition state mimetic (WEHI-842) | | Descriptor: | (2R)-1-[(2R)-2-(2-methoxyethoxy)propoxy]propan-2-amine, 1,2-ETHANEDIOL, Aspartic protease PM5, ... | | Authors: | Czabotar, P.E, Hodder, A.N, Smith, B.J, Sleebs, B.E, Gazdic, M, Boddey, J.A, Cowman, A.F. | | Deposit date: | 2015-05-01 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis for plasmepsin V inhibition that blocks export of malaria proteins to human erythrocytes.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5T20
 
 | | Crystal Structure of Tarin Lectin bound to Trimannose | | Descriptor: | 1,2-ETHANEDIOL, Lectin, alpha-D-mannopyranose, ... | | Authors: | Pereira, P.R, Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2016-08-22 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | High-resolution crystal structures of Colocasia esculenta tarin lectin.
Glycobiology, 27, 2017
|
|
4WUV
 
 | | Crystal Structure of a putative D-Mannonate oxidoreductase from Haemophilus influenza (Avi_5165, TARGET EFI-513796) with bound NAD | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2014-11-03 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of a putative D-Mannonate oxidoreductase from Haemophilus influenza (Avi_5165, TARGET EFI-513796) with bound NAD
To be published
|
|
4WXE
 
 | | CRYSTAL STRUCTURE OF A LACI REGULATOR FROM LACTOBACILLUS CASEI (LSEI_2103, TARGET EFI-512911) WITH BOUND TRIS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-13 | | Release date: | 2014-11-26 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF A LACI REGULATOR FROM LACTOBACILLUS CASEI (LSEI_2103, TARGET EFI-512911) WITH BOUND TRIS
To be published
|
|
5V4L
 
 | |
3UPH
 
 | | Synthesis of novel 4,5-dihydrofurano indoles and their evaluation as HCV NS5B polymerase inhibitors | | Descriptor: | 6-(2,5-difluorobenzyl)-N-(methylsulfonyl)-8-(2-oxo-1,2-dihydropyridin-3-yl)-3,6-dihydro-2H-furo[2,3-e]indole-7-carboxamide, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Velazquez, F, Venkataraman, S, Lesburg, C.A, Duca, J.S, Rosenblum, S.B, Kozlowski, J.A, Njoroge, F.G. | | Deposit date: | 2011-11-18 | | Release date: | 2012-01-25 | | Last modified: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis of New 4,5-Dihydrofuranoindoles and Their Evaluation as HCV NS5B Polymerase Inhibitors.
Org.Lett., 14, 2012
|
|
4X08
 
 | | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms. | | Descriptor: | Eosinophil cationic protein, SULFATE ION | | Authors: | Blanco, J.A, Garcia, J.M, Salazar, V.A, Sanchez, D, Moussauoi, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure of H128N/ECP mutant in complex with sulphate anions at 1.34 Angstroms.
To Be Published
|
|
3USR
 
 | | Structure of Y194F glycogenin mutant truncated at residue 270 | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycogenin-1 | | Authors: | Issoglio, F.M, Carrizo, M.E, Romero, J.M, Curtino, J.A. | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanisms of monomeric and dimeric glycogenin autoglucosylation.
J.Biol.Chem., 287, 2012
|
|
4YZ1
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, apo structure. | | Descriptor: | Putative neuraminidase, SULFATE ION | | Authors: | Lukacik, P, Owen, D.O, Potter, J.A, Taylor, G.L, Walsh, M.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
4Z0N
 
 | | Crystal Structure of a Periplasmic Solute binding protein (IPR025997) from Streptobacillus moniliformis DSM-12112 (Smon_0317, TARGET EFI-511281) with bound D-Galactose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal Structure of a Periplasmic Solute binding protein (IPR025997) from Streptobacillus moniliformis DSM-12112 (Smon_0317, TARGET EFI-511281) with bound D-Galactose
To be published
|
|
4YZ2
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, in complex with 2-deoxy-2,3-didehydro-N-acetylneuraminic acid. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, SULFATE ION, Sialidase NanC | | Authors: | Lukacik, P, Owen, C.D, Potter, J.A, Taylor, G.L, Walsh, M.A. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
4Z93
 
 | | BRD4 bromodomain 2 in complex with gamma-carboline-containing compound, number 18. | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-cyclopropyl-5-methyl-1H-pyrazol-4-yl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-8-methoxy-5H-pyrido[4,3-b]indole, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2015-04-09 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structure-Based Design of gamma-Carboline Analogues as Potent and Specific BET Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
4YZV
 
 | | Precleavage 70S structure of the P. vulgaris HigB deltaH92 toxin bound to the ACA codon | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Schureck, M.A, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2015-03-25 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Defining the mRNA recognition signature of a bacterial toxin protein.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5UP9
 
 | | Crystal Structure of Zn-bound Human Heavy-Chain ferritin variant 122H-delta C-star with para-xylenedihydroxamate | | Descriptor: | 2,2'-(1,4-phenylene)bis(N-hydroxyacetamide), DI(HYDROXYETHYL)ETHER, Ferritin heavy chain, ... | | Authors: | Bailey, J.B, Zhang, L, Chiong, J.A, Ahn, S, Tezcan, F.A. | | Deposit date: | 2017-02-01 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthetic Modularity of Protein-Metal-Organic Frameworks.
J. Am. Chem. Soc., 139, 2017
|
|
5V5C
 
 | | VQIINK, Structure of the amyloid-spine from microtubule associated protein tau Repeat 2 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Seidler, P.M, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S, Cascio, D, Boyer, D.R. | | Deposit date: | 2017-03-14 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.25 Å) | | Cite: | Structure-based inhibitors of tau aggregation.
Nat Chem, 10, 2018
|
|
446D
 
 | | STRUCTURE OF THE OLIGONUCLEOTIDE D(CGTATATACG) AS A SITE SPECIFIC COMPLEX WITH NICKEL IONS | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*TP*AP*TP*AP*CP*G)-3'), NICKEL (II) ION | | Authors: | Abrescia, N.G.A, Malinina, L, Gonzaga, L.F, Huynh-Dinh, T, Neidle, S, Subirana, J.A. | | Deposit date: | 1999-01-15 | | Release date: | 1999-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the oligonucleotide d(CGTATATACG) as a site-specific complex with nickel ions.
Nucleic Acids Res., 27, 1999
|
|
4WWH
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEG_1704, TARGET EFI-510967) WITH BOUND D-GALACTOSE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, beta-D-galactopyranose | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEG_1704, TARGET EFI-510967) WITH BOUND D-GALACTOSE
To be published
|
|
4WYQ
 
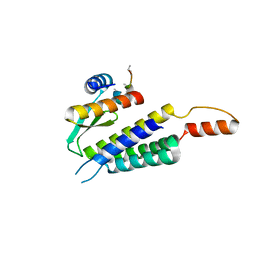 | | Crystal structure of the Dicer-TRBP interface | | Descriptor: | Endoribonuclease Dicer, Poly(UNK), RISC-loading complex subunit TARBP2 | | Authors: | Wilson, R.C, Doudna, J.A. | | Deposit date: | 2014-11-18 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dicer-TRBP Complex Formation Ensures Accurate Mammalian MicroRNA Biogenesis.
Mol.Cell, 57, 2015
|
|
5UU5
 
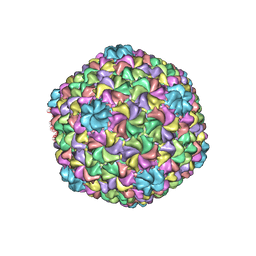 | | Bacteriophage P22 mature virion capsid protein | | Descriptor: | Major capsid protein | | Authors: | Hryc, C.F, Chen, D.-H, Afonine, P.V, Jakana, J, Wang, Z, Haase-Pettingell, C, Jiang, W, Adams, P.D, King, J.A, Schmid, M.F, Chiu, W. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Accurate model annotation of a near-atomic resolution cryo-EM map.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UX0
 
 | |
4A8L
 
 | | Protein crystallization and microgravity: glucose isomerase crystals grown during the PCDF-PROTEIN mission | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, XYLOSE ISOMERASE | | Authors: | Decanniere, K, Patino-Lopez, L.-D, Sleutel, M, Evrard, C, Van De Weerdt, C, Haumont, E, Gavira, J.A, Otalora, F, Maes, D. | | Deposit date: | 2011-11-21 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Protein Crystallization and Microgravity: Glucose Isomerase Crystals Grown During the Pcdf-Protein Mission
To be Published
|
|
4AJN
 
 | | rat LDHA in complex with 2-((4-(2-((3-((2-methyl-1,3-benzothiazol-6- yl)amino)-3-oxo-propyl)carbamoylamino)ethyl)phenyl)methyl) propanedioic acid | | Descriptor: | (4-{2-[({3-[(2-METHYL-1,3-BENZOTHIAZOL-6-YL)AMINO]-3-OXOPROPYL}CARBAMOYL)AMINO]ETHYL}BENZYL)PROPANEDIOIC ACID, GLYCEROL, L-LACTATE DEHYDROGENASE A CHAIN | | Authors: | Tucker, J.A, Brassington, C, Hassall, G, Watson, M, Ward, R, Tart, J, Davies, G, Caputo, A, Pearson, S. | | Deposit date: | 2012-02-16 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Design and Synthesis of Novel Lactate Dehydrogenase a Inhibitors by Fragment-Based Lead Generation
J.Med.Chem., 55, 2012
|
|
