4V6E
 
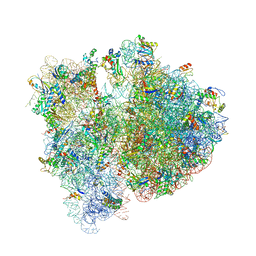 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.712 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
4UUC
 
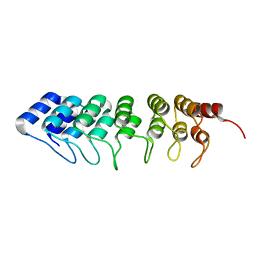 | | Crystal structure of human ASB11 ankyrin repeat domain | | Descriptor: | ANKYRIN REPEAT AND SOCS BOX PROTEIN 11 | | Authors: | Pinkas, D.M, Sanvitale, C, Kragh Nielsen, T, Guo, K, Sorrell, F, Berridge, G, Ayinampudi, V, Wang, D, Newman, J.A, Tallant, C, Chaikuad, A, Canning, P, Kopec, J, Krojer, T, Vollmar, M, Allerston, C.K, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Bullock, A. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Asb11 Ankyrin Repeat Domain
To be Published
|
|
8RHF
 
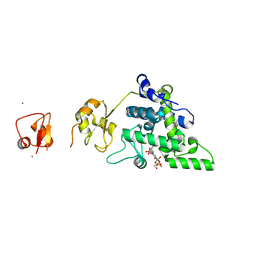 | | Lytic Transglycosylase MltD of Pseudomonas aeruginosa bound to the Natural Product Bulgecin A, with two LysM domains | | Descriptor: | 4-O-(4-O-SULFONYL-N-ACETYLGLUCOSAMININYL)-5-METHYLHYDROXY-L-PROLINE-TAURINE, DI(HYDROXYETHYL)ETHER, LysM peptidoglycan-binding domain-containing protein, ... | | Authors: | Miguel-Ruano, V, Hermoso, J.A. | | Deposit date: | 2023-12-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of lytic transglycosylase MltD of Pseudomonas aeruginosa, a target for the natural product bulgecin A.
Int.J.Biol.Macromol., 267, 2024
|
|
3S3B
 
 | | Structure of Thermus thermophilus cytochrome ba3 oxidase 240s after Xe depressurization | | Descriptor: | COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Luna, V.M, Fee, J.A, Deniz, A.A, Stout, C.D. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mobility of Xe atoms within the oxygen diffusion channel of cytochrome ba(3) oxidase.
Biochemistry, 51, 2012
|
|
249D
 
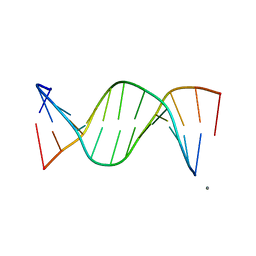 | | STRUCTURAL COMPARISON BETWEEN THE D(CTAG) SEQUENCE IN OLIGONUCLEOTIDES AND TRP AND MET REPRESSOR-OPERATOR COMPLEXES | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*GP*CP*TP*CP*TP*AP*GP*AP*GP*CP*G)-3') | | Authors: | Urpi, L, Tereshko, V, Malinina, L, Huynh-Dinh, T, Subirana, J.A. | | Deposit date: | 1996-02-22 | | Release date: | 1996-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural comparison between the d(CTAG) sequence in oligonucleotides and trp and met repressor-operator complexes.
Nat.Struct.Biol., 3, 1996
|
|
5CFL
 
 | | Crystal structure of anemone STING (Nematostella vectensis) in complex with 3', 3' c-di-GMP, c[G(3', 5')pG(3', 5')p] | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CITRATE ANION, Stimulator of Interferon Genes | | Authors: | Kranzusch, P.J, Wilson, S.C, Lee, A.S.Y, Berger, J.M, Doudna, J.A, Vance, R.E. | | Deposit date: | 2015-07-08 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.836 Å) | | Cite: | Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2',3' cGAMP Signaling.
Mol.Cell, 59, 2015
|
|
3S39
 
 | | Structure of Thermus thermophilus cytochrome ba3 oxidase 60s after Xe depressurization | | Descriptor: | COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Luna, V.M, Fee, J.A, Deniz, A.A, Stout, C.D. | | Deposit date: | 2011-05-17 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Mobility of Xe atoms within the oxygen diffusion channel of cytochrome ba(3) oxidase.
Biochemistry, 51, 2012
|
|
8GHS
 
 | | Empty HBV Cp183 capsid with importin-beta, subparticle reconstruction at 2-fold location | | Descriptor: | Capsid protein | | Authors: | Kim, C, Schlicksup, C.J, Hadden-Perilla, J.A, Wang, J.C.-Y, Zlotnick, A. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the Hepatitis B virus capsid quasi-6-fold with a trapped C-terminal domain reveals capsid movements associated with domain exit.
J.Biol.Chem., 299, 2023
|
|
5CH7
 
 | | Crystal structure of the perchlorate reductase PcrAB - Phe164 gate switch intermediate - from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ACETATE ION, ... | | Authors: | Tsai, C.-L, Tainer, J.A. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
4DBE
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Sulfolobus solfataricus complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-01-14 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Sulfolobus solfataricus complexed with inhibitor BMP
To be Published
|
|
4UTW
 
 | | Structural characterisation of NanE, ManNac6P C2 epimerase, from Clostridium perfingens | | Descriptor: | CHLORIDE ION, N-acetyl-D-glucosamine-6-phosphate, PUTATIVE N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
4DX3
 
 | | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue | | Descriptor: | CHLORIDE ION, GLYCEROL, Mandelate racemase / muconate lactonizing enzyme family protein | | Authors: | Vetting, M.W, Bouvier, J.T, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-27 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup, target EFI-502086) from Agrobacterium tumefaciens, with a succinimide residue
to be published
|
|
4UVH
 
 | | Discovery of pyrimidine isoxazoles InhA in complex with compound 10 | | Descriptor: | ACETATE ION, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], N-(1,3-BENZOTHIAZOL-2-YL)ACETAMIDE, ... | | Authors: | Read, J.A, Gingell, H, Madhavapeddi, P, Ghorpade, S, Cowan, S. | | Deposit date: | 2014-08-05 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Hitting the Target in More Than One Way: Novel, Direct Inhibitors of Mycobacterium Tuberculosis Enoyl Acp Reductase
To be Published
|
|
5CM6
 
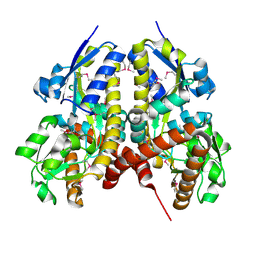 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM PSEUDOALTEROMONAS ATLANTICA T6c(Patl_2292, TARGET EFI-510180) WITH BOUND SODIUM AND PYRUVATE | | Descriptor: | PYRUVIC ACID, SODIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-07-16 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM PSEUDOALTEROMONAS ATLANTICA T6c(Patl_2292, TARGET EFI-510180) WITH BOUND SODIUM AND PYRUVATE
To be published
|
|
4UWJ
 
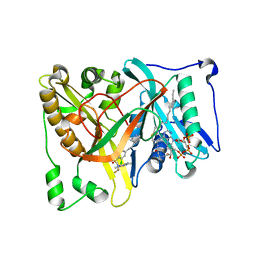 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a capped pyrazole sulphonamide ligand | | Descriptor: | 2,6-dichloro-N-(difluoromethyl)-4-[3-(piperidin-4-yl)propyl]-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Norcross, N.R, Thompson, S, Harrison, J.R, Smith, V.C, Torrie, L.S, McElroy, S.P, Hallyburton, I, Norval, S, Stojanovski, L, Simeons, F.R.C, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lead optimization of a pyrazole sulfonamide series of Trypanosoma brucei N-myristoyltransferase inhibitors: identification and evaluation of CNS penetrant compounds as potential treatments for stage 2 human African trypanosomiasis.
J. Med. Chem., 57, 2014
|
|
4V7T
 
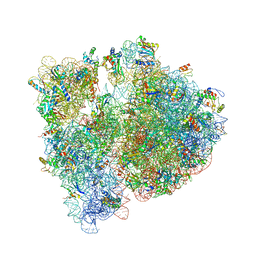 | | Crystal structure of the E. coli ribosome bound to chloramphenicol. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-14 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1942 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5CNJ
 
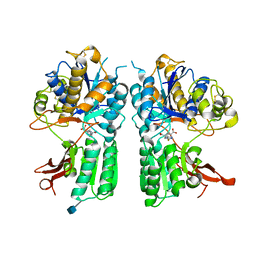 | | mGlur2 with glutamate analog | | Descriptor: | (1R,2S,4R,5R,6R)-2-amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | Authors: | Monn, J.A, Clawson, D.K. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-(Thiotriazolyl)-substituted-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1R,2S,4R,5R,6R)-2-Amino-4-(1H-1,2,4-triazol-3-ylsulfanyl)bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2812223), a Highly Potent, Functionally Selective mGlu2 Receptor Agonist.
J.Med.Chem., 58, 2015
|
|
4UWI
 
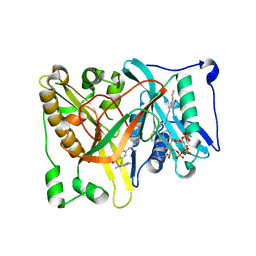 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a pyrazole sulphonamide ligand | | Descriptor: | 2,6-dichloro-4-[3-(4-methylpiperazin-1-yl)propyl]-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Norcross, N.R, Thompson, S, Harrison, J.R, Smith, V.C, Torrie, L.S, McElroy, S.P, Hallyburton, I, Norval, S, Stojanovski, L, Simeons, F.R.C, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lead optimization of a pyrazole sulfonamide series of Trypanosoma brucei N-myristoyltransferase inhibitors: identification and evaluation of CNS penetrant compounds as potential treatments for stage 2 human African trypanosomiasis.
J. Med. Chem., 57, 2014
|
|
4V97
 
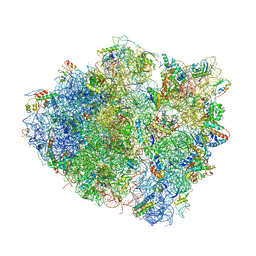 | | Crystal structure of the bacterial ribosome ram mutation G299A. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fagan, C.E, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2012-04-06 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.516 Å) | | Cite: | Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8GRJ
 
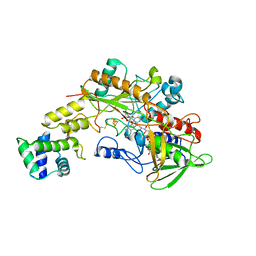 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yoshida, H, Kojima, K, Tsugawa, W, Okuda-Shimazaki, J, Kerrigan, J.A, Sode, K. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone
To Be Published
|
|
8GJ9
 
 | |
8GJA
 
 | | RAD51C-XRCC3 structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, RAD51C, ... | | Authors: | Arvai, A.S, Tainer, J.A, Williams, G, Longo, M.A. | | Deposit date: | 2023-03-15 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
8G2V
 
 | | Cryo-EM structure of recombinant human LECT2 amyloid fibril core | | Descriptor: | Leukocyte cell-derived chemotaxin-2 | | Authors: | Richards, L.S, Flores, M.D, Zink, S, Schibrowsky, N.A, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2023-02-06 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.715 Å) | | Cite: | Cryo-EM structure of a human LECT2 amyloid fibril reveals a network of polar ladders at its core.
Structure, 31, 2023
|
|
4V0C
 
 | |
5CGC
 
 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, ... | | Authors: | Christopher, J.A, Aves, S.J, Bennett, K.A, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Okrasa, K, Serrano-Vega, M.J, Tehan, B.G, Wiggin, G.R, Congreve, M. | | Deposit date: | 2015-07-09 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile).
J.Med.Chem., 58, 2015
|
|
