8AMV
 
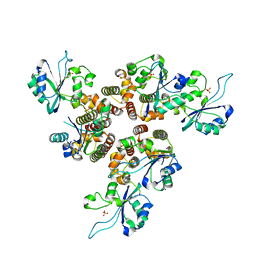 | | RepB pMV158 hexamer | | Descriptor: | PHOSPHATE ION, Replication protein RepB, SODIUM ION | | Authors: | Machon, C, Amodio, J, Boer, R.D, Ruiz-Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structures of pMV158 replication initiator RepB with and without DNA reveal a flexible dual-function protein.
Nucleic Acids Res., 51, 2023
|
|
8AMU
 
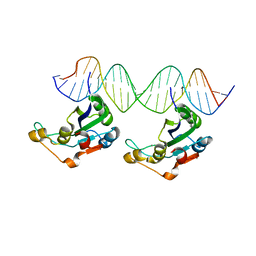 | | RepB pMV158 OBD domain bound to DDR region | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*GP*TP*CP*GP*CP*CP*GP*AP*AP*AP*AP*GP*TP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*AP*CP*TP*TP*TP*TP*CP*GP*GP*CP*GP*AP*CP*TP*TP*TP*T)-3'), MANGANESE (II) ION, ... | | Authors: | Amodio, J, Machon, C, Boer, R.D, Ruiz-Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2022-08-04 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of pMV158 replication initiator RepB with and without DNA reveal a flexible dual-function protein.
Nucleic Acids Res., 51, 2023
|
|
6TAT
 
 | |
7BLO
 
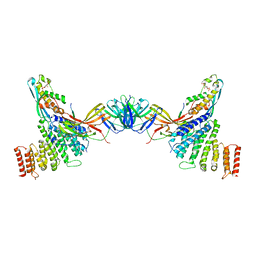 | | VPS26 dimer region of metazoan membrane-assembled retromer:SNX3 complex modelled with human proteins | | Descriptor: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, C-term (residues 493-54) of Wls (fitted sequence corresponds to hDMT1-II), Sorting nexin-3, ... | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-03 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
6FPP
 
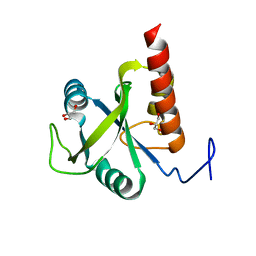 | | Structure of S. pombe Mmi1 | | Descriptor: | GLYCEROL, MAGNESIUM ION, YTH domain-containing protein mmi1 | | Authors: | Stowell, J.A.W, Hill, C.H, Yu, M, Wagstaff, J.L, McLaughlin, S.H, Freund, S.M.V, Passmore, L.A. | | Deposit date: | 2018-02-11 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A low-complexity region in the YTH domain protein Mmi1 enhances RNA binding.
J. Biol. Chem., 293, 2018
|
|
5KZE
 
 | | N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus | | Descriptor: | GLYCEROL, N-acetylneuraminate lyase, SULFATE ION | | Authors: | North, R.A, Watson, A.J.A, Pearce, F.G, Muscroft-Taylor, A.C, Friemann, R, Fairbanks, A.J, Dobson, R.C.J. | | Deposit date: | 2016-07-25 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure and inhibition of N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus.
FEBS Lett., 590, 2016
|
|
6TB8
 
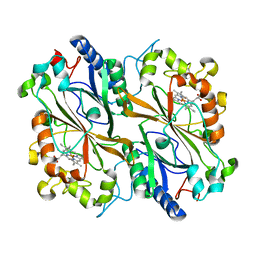 | | Dye Type Peroxidase Aa from Streptomyces lividans: spectroscopically-validated ferric state | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lucic, M, Moreno-Chicano, T.M, Hough, M.A, Dworkowski, F.S.N, Worrall, J.A.R. | | Deposit date: | 2019-11-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A subtle structural change in the distal haem pocket has a remarkable effect on tuning hydrogen peroxide reactivity in dye decolourising peroxidases from Streptomyces lividans.
Dalton Trans, 49, 2020
|
|
7BL1
 
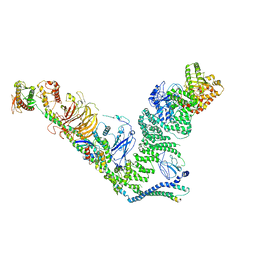 | | human complex II-BATS bound to membrane-attached Rab5a-GTP | | Descriptor: | Beclin-1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tremel, S, Morado, D.R, Kovtun, O, Williams, R.L, Briggs, J.A.G, Munro, S, Ohashi, Y, Bertram, J, Perisic, O. | | Deposit date: | 2021-01-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes.
Nat Commun, 12, 2021
|
|
5NLB
 
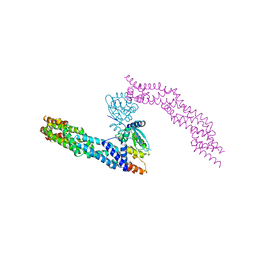 | | Crystal structure of human CUL3 N-terminal domain bound to KEAP1 BTB and 3-box | | Descriptor: | Cullin-3, Kelch-like ECH-associated protein 1 | | Authors: | Adamson, R, Krojer, T, Pinkas, D.M, Bartual, S.G, Burgess-Brown, N.A, Borkowska, O, Chalk, R, Newman, J.A, Kopec, J, Dixon-Clarke, S.E, Mathea, S, Sethi, R, Velupillai, S, Mackinnon, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2017-04-04 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural and biochemical characterization establishes a detailed understanding of KEAP1-CUL3 complex assembly.
Free Radic Biol Med, 204, 2023
|
|
7BLQ
 
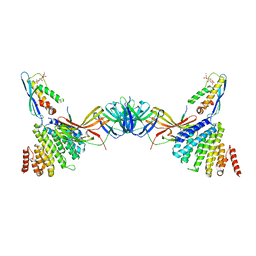 | | Vps26 dimer region of the fungal membrane-assembled retromer:Grd19 complex. | | Descriptor: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, Sorting nexin-3, The C-terminal portion of Kex2 cargo, ... | | Authors: | Leneva, N, Kovtun, O, Morado, D.R, Briggs, J.A.G, Owen, D.J. | | Deposit date: | 2021-01-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly.
Sci Adv, 7, 2021
|
|
1ISC
 
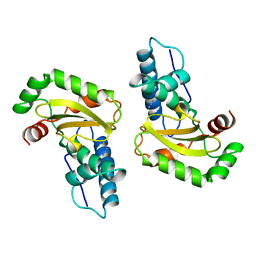 | | STRUCTURE-FUNCTION IN E. COLI IRON SUPEROXIDE DISMUTASE: COMPARISONS WITH THE MANGANESE ENZYME FROM T. THERMOPHILUS | | Descriptor: | AZIDE ION, FE (III) ION, IRON(III) SUPEROXIDE DISMUTASE | | Authors: | Lah, M.S, Dixon, M, Pattridge, K.A, Stallings, W.C, Fee, J.A, Ludwig, M.L. | | Deposit date: | 1994-07-12 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus.
Biochemistry, 34, 1995
|
|
6T63
 
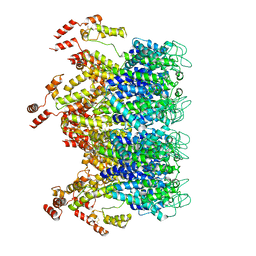 | | A model of the EIAV CA-SP hexamer (C2) from Gag-deltaMA tubes assembled at pH6 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
1QS2
 
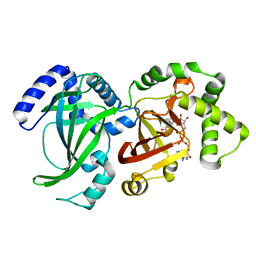 | | CRYSTAL STRUCTURE OF VIP2 WITH NAD | | Descriptor: | ADP-RIBOSYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Han, S, Craig, J.A, Putnam, C.D, Carozzi, N.B, Tainer, J.A. | | Deposit date: | 1999-06-25 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex.
Nat.Struct.Biol., 6, 1999
|
|
8ACT
 
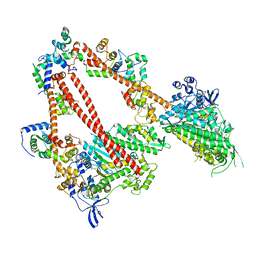 | | structure of the human beta-cardiac myosin folded-back off state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin light chain 3, ... | | Authors: | Grinzato, A, Kandiah, E, Robert-Paganin, J, Auguin, D, Kikuti, C, Nandwani, N, Moussaoui, D, Pathak, D, Ruppel, K.M, Spudich, J.A, Houdusse, A. | | Deposit date: | 2022-07-06 | | Release date: | 2023-06-07 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the folded-back state of human beta-cardiac myosin.
Nat Commun, 14, 2023
|
|
6TAP
 
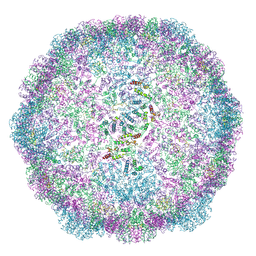 | | Structure of the dArc1 capsid | | Descriptor: | Activity-regulated cytoskeleton associated protein 1, ZINC ION | | Authors: | Erlendsson, S, Morado, D.R, Shepherd, J.D, Briggs, J.A.G. | | Deposit date: | 2019-10-30 | | Release date: | 2020-01-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of virus-like capsids formed by the Drosophila neuronal Arc proteins.
Nat.Neurosci., 23, 2020
|
|
8S9L
 
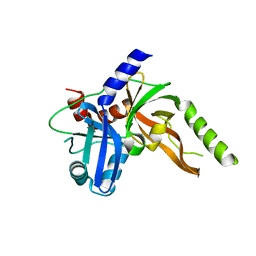 | | Structure of monomeric FAM111A SPD V347D Mutant | | Descriptor: | SULFATE ION, Serine protease FAM111A | | Authors: | Palani, S, Alvey, J.A, Cong, A.T.Q, Schellenberg, M.J, Machida, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes.
Nat Commun, 15, 2024
|
|
8S9K
 
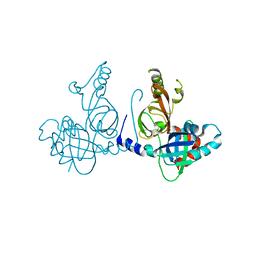 | | Structure of dimeric FAM111A SPD S541A Mutant | | Descriptor: | GLYCEROL, Serine protease FAM111A | | Authors: | Palani, S, Alvey, J.A, Cong, A.T.Q, Schellenberg, M.J, Machida, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Dimerization-dependent serine protease activity of FAM111A prevents replication fork stalling at topoisomerase 1 cleavage complexes.
Nat Commun, 15, 2024
|
|
1QS1
 
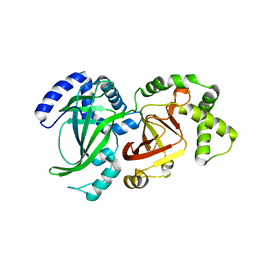 | | CRYSTAL STRUCTURE OF VEGETATIVE INSECTICIDAL PROTEIN2 (VIP2) | | Descriptor: | ADP-RIBOSYLTRANSFERASE | | Authors: | Han, S, Craig, J.A, Putnam, C.D, Carozzi, N.B, Tainer, J.A. | | Deposit date: | 1999-06-25 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex.
Nat.Struct.Biol., 6, 1999
|
|
5N97
 
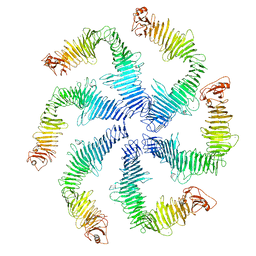 | | Structure of the C. crescentus S-layer | | Descriptor: | CALCIUM ION, S-layer protein rsaA | | Authors: | Bharat, T.A, Hagen, W.J, Briggs, J.A, Lowe, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the hexagonal surface layer on Caulobacter crescentus cells.
Nat Microbiol, 2, 2017
|
|
5H8H
 
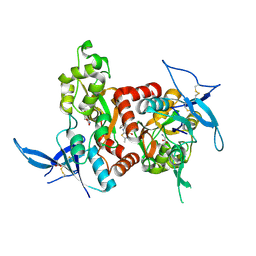 | | Structure of the human GluN1/GluN2A LBD in complex with GNE3419 | | Descriptor: | 7-[[ethyl(phenyl)amino]methyl]-2-methyl-[1,3,4]thiadiazolo[3,2-a]pyrimidin-5-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function.
Neuron, 89, 2016
|
|
6FPX
 
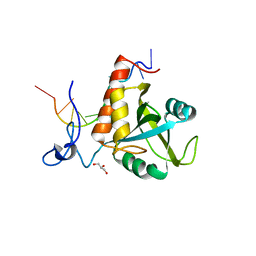 | | Structure of S. pombe Mmi1 in complex with 11-mer RNA | | Descriptor: | GLYCEROL, RNA (5'-R(P*UP*UP*UP*AP*AP*AP*CP*CP*UP*A)-3'), YTH domain-containing protein mmi1 | | Authors: | Stowell, J.A.W, Hill, C.H, Yu, M, Wagstaff, J.L, McLaughlin, S.H, Freund, S.M.V, Passmore, L.A. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A low-complexity region in the YTH domain protein Mmi1 enhances RNA binding.
J. Biol. Chem., 293, 2018
|
|
6T61
 
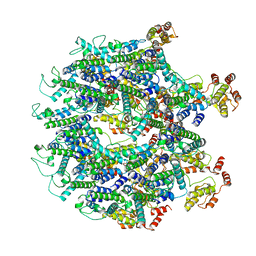 | | A model of the EIAV CA-SP hexamer (C2) from Gag-deltaMA tubes assembled at pH8 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
2XMH
 
 | | The X-ray structure of CTP:inositol-1-phosphate cytidylyltransferase from Archaeoglobus fulgidus | | Descriptor: | CITRATE ANION, CTP-INOSITOL-1-PHOSPHATE CYTIDYLYLTRANSFERASE | | Authors: | Brito, J.A, Borges, N, Vonrhein, C, Santos, H, Archer, M. | | Deposit date: | 2010-07-27 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Archaeoglobus fulgidus CTP:inositol-1-phosphate cytidylyltransferase, a key enzyme for di-myo-inositol-phosphate synthesis in (hyper)thermophiles.
J. Bacteriol., 193, 2011
|
|
8AGS
 
 | | Cyclohexane epoxide soak of epoxide hydrolase from metagenomic source ch65 resulting in halogenated compound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 2-CHLOROPHENOL, Alpha/beta epoxide hydrolase, ... | | Authors: | Isupov, M.N, De Rose, S.A, Mitchell, D, Littlechild, J.A. | | Deposit date: | 2022-07-20 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Complexes of epoxide hydrolase from metagenomic source ch65
To Be Published
|
|
3PTE
 
 | |
