4PEM
 
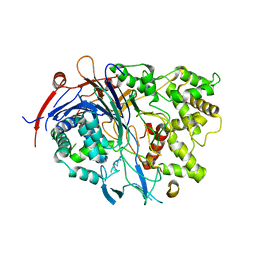 | | Crystal Structure of S1G mutant of Penicillin G Acylase from Kluyvera citrophila | | Descriptor: | CALCIUM ION, Penicillin G acylase Alpha, Penicillin G acylase Beta | | Authors: | Ramasamy, S, Varshney, N.K, Brannigan, J.A, Wilkinson, A.J, Suresh, C.G. | | Deposit date: | 2014-04-24 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of S1G mutant of Penicillin G Acylase from Kluyvera citrophilla
To be Published
|
|
4PF8
 
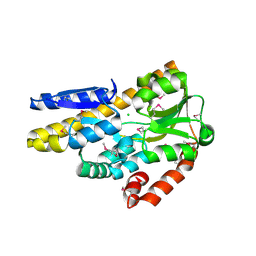 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM SULFITOBACTER sp. NAS-14.1 (TARGET EFI-510299) WITH BOUND BETA-D-GALACTURONATE | | Descriptor: | CHLORIDE ION, TRAP-T family transporter, DctP (Periplasmic binding) subunit, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MSQ
 
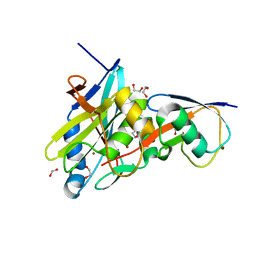 | | Crystal structure of Schizosaccharomyces pombe AMSH-like protease sst2 catalytic domain bound to ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease sst2, PHOSPHATE ION, ... | | Authors: | Shrestha, R.K, Ronau, J.A, Das, C. | | Deposit date: | 2013-09-18 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Insights into the Mechanism of Deubiquitination by JAMM Deubiquitinases from Cocrystal Structures of the Enzyme with the Substrate and Product.
Biochemistry, 53, 2014
|
|
3E4R
 
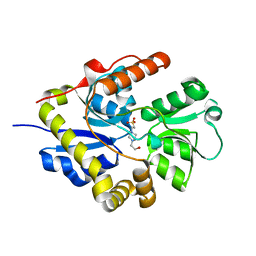 | | Crystal structure of the alkanesulfonate binding protein (SsuA) from the phytopathogenic bacteria Xanthomonas axonopodis pv. citri bound to HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nitrate transport protein | | Authors: | Balan, A, Araujo, F.T, Sanches, M, Chirgadze, D.Y, Blundell, T.B, Barbosa, J.A.R.G. | | Deposit date: | 2008-08-12 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the alkanesulfonate binding protein (SsuA) from the phytopathogenic bacteria Xanthomonas axonopodis pv. citri bound to HEPES
To be Published
|
|
4MZL
 
 | | Crystal Structure of MTIP from Plasmodium falciparum in complex with HBS myoA, a hydrogen bond surrogate myoA helix mimetic | | Descriptor: | Myosin A tail domain interacting protein, hydrogen bond surrogate (HBS) myoA helix mimetic | | Authors: | Douse, C.H, Garnett, J.A, Maas, S.J, Cota, E, Tate, E.W. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structures of Stapled and Hydrogen Bond Surrogate Peptides Targeting a Fully Buried Protein-Helix Interaction.
Acs Chem.Biol., 8, 2014
|
|
4MPF
 
 | | Crystal structure of human glutathione transferase theta-2, complex with inorganic phosphate, GSH free, target EFI-507257 | | Descriptor: | Glutathione S-transferase theta-2, L(+)-TARTARIC ACID, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Glutathione S-Transferase Theta-2 (Target Efi-507257)
To be Published
|
|
4N8Y
 
 | | Crystal structure of a trap periplasmic solute binding protein from bradyrhizobium sp. btai1 b (bbta_0128), target EFI-510056 (bbta_0128), complex with alpha/beta-d-galacturonate | | Descriptor: | Putative TRAP-type C4-dicarboxylate transport system, binding periplasmic protein (DctP subunit), alpha-D-galactopyranuronic acid, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MZJ
 
 | | Crystal Structure of MTIP from Plasmodium falciparum in complex with pGly[801,805], a stapled myoA tail peptide | | Descriptor: | Myosin A tail domain interacting protein, Myosin-A | | Authors: | Douse, C.H, Garnett, J.A, Maas, S.J, Cota, E, Tate, E.W. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | Crystal Structures of Stapled and Hydrogen Bond Surrogate Peptides Targeting a Fully Buried Protein-Helix Interaction.
Acs Chem.Biol., 8, 2014
|
|
6GIL
 
 | | NMR structure of temporin B in SDS micelles | | Descriptor: | Temporin-B | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
6GS5
 
 | | NMR structure of temporin L in SDS micelles | | Descriptor: | Temporin-L | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-06-13 | | Release date: | 2018-07-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Temporin L and aurein 2.5 have identical conformations but subtly distinct membrane and antibacterial activities.
Sci Rep, 9, 2019
|
|
6GS9
 
 | | NMR structure of aurein 2.5 in SDS micelles | | Descriptor: | Aurein 2.5 | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-06-13 | | Release date: | 2018-07-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Temporin L and aurein 2.5 have identical conformations but subtly distinct membrane and antibacterial activities.
Sci Rep, 9, 2019
|
|
6GIJ
 
 | | NMR structure of temporin B KKG6A in SDS micelles | | Descriptor: | temporinB_KKG6A | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
4O94
 
 | | Crystal structure of a trap periplasmic solute binding protein from Rhodopseudomonas palustris HaA2 (RPB_3329), Target EFI-510223, with bound succinate | | Descriptor: | CHLORIDE ION, SUCCINIC ACID, TRAP dicarboxylate transporter DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-01-01 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4O7M
 
 | | Crystal structure of a trap periplasmic solute binding protein from shewanella loihica PV-4, target EFI-510273, with bound L-malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-25 | | Release date: | 2014-03-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PG1
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | (1S,2S)-2-nonylcyclopropanecarboxylic acid, Aldehyde decarbonylase, DIMETHYL SULFOXIDE, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-05-01 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4PGI
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Analogs Bound | | Descriptor: | 1,2-ETHANEDIOL, 11-[2-(2-ethoxyethoxy)ethoxy]undecanal, Aldehyde decarbonylase, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-05-02 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4PPE
 
 | | human RNF4 RING domain | | Descriptor: | E3 ubiquitin-protein ligase RNF4, ZINC ION | | Authors: | Perry, J.J, Arvai, A.S, Hitomi, C, Tainer, J.A. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RNF4 interacts with both SUMO and nucleosomes to promote the DNA damage response.
Embo Rep., 15, 2014
|
|
4PRF
 
 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | Descriptor: | Hepatitis Delta virus ribozyme, STRONTIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
4PRU
 
 | | Crystal structure of dimethyllysine hen egg-white lysozyme in complex with sclx4 at 2.2 A resolution | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, GLYCEROL, Lysozyme C | | Authors: | McGovern, R.E, Lyons, J.A, Crowley, P.B. | | Deposit date: | 2014-03-06 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural study of a small molecule receptor bound to dimethyllysine in lysozyme.
Chem Sci, 6, 2015
|
|
4PTS
 
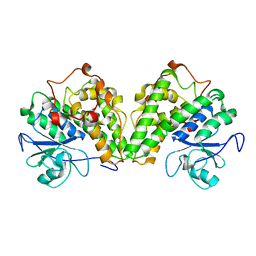 | | Crystal structure of a glutathione transferase from Gordonia bronchialis DSM 43247, target EFI-507405 | | Descriptor: | glutathione S-transferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-11 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of a glutathione transferase from Gordonia bronchialis DSM 43247, target EFI-507405
To be Published
|
|
4PFB
 
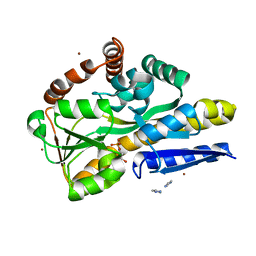 | | Crystal structure of a TRAP periplasmic solute binding protein from Fusobacterium nucleatun (FN1258, TARGET EFI-510120) with bound SN-glycerol-3-phosphate | | Descriptor: | C4-dicarboxylate-binding protein, IMIDAZOLE, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MIJ
 
 | | Crystal structure of a Trap periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Galacturonate, space group P21 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-31 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MMW
 
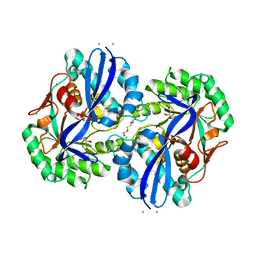 | | Crystal structure of D-glucarate dehydratase from Agrobacterium tumefaciens complexed with magnesium, L-Xylarohydroxamate and L-Lyxarohydroxamate | | Descriptor: | (2R,3S,4R)-2,3,4-TRIHYDROXY-5-(HYDROXYAMINO)-5-OXOPENTANOIC ACID, CHLORIDE ION, Isomerase/lactonizing enzyme, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-09-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal structure of D-glucarate dehydratase from Agrobacterium tumefaciens complexed with magnesium, L-Xylarohydroxamate and L-Lyxarohydroxamate
To be Published
|
|
4MNI
 
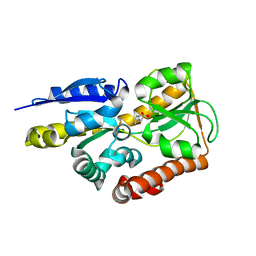 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_4736), Target EFI-510156, with bound benzoyl formate, space group P6522 | | Descriptor: | BENZOYL-FORMIC ACID, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MSD
 
 | | Crystal structure of Schizosaccharomyces pombe AMSH-like protein SST2 T319I mutant | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, AMSH-like protease sst2, ... | | Authors: | Shrestha, R.K, Ronau, J.A, Das, C. | | Deposit date: | 2013-09-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Mechanism of Deubiquitination by JAMM Deubiquitinases from Cocrystal Structures of the Enzyme with the Substrate and Product.
Biochemistry, 53, 2014
|
|
