5D55
 
 | | Crystal structure of the E. coli Hda pilus minor tip subunit, HdaB | | Descriptor: | CITRATE ANION, HdaB,HdaA (Adhesin), HUS-associated diffuse adherence, ... | | Authors: | Lee, W.-C, Garnett, J.A, Matthews, S.J. | | Deposit date: | 2015-08-10 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and analysis of HdaB: The enteroaggregative Escherichia coli AAF/IV pilus tip protein.
Protein Sci., 25, 2016
|
|
2BZB
 
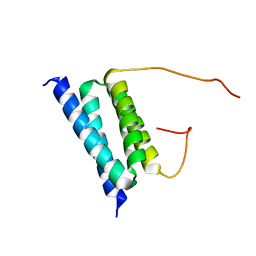 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-08-14 | | Release date: | 2006-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
2BW2
 
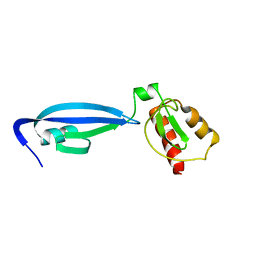 | | BofC from Bacillus subtilis | | Descriptor: | BYPASS OF FORESPORE C | | Authors: | Patterson, H.M, Brannigan, J.A, Cutting, S.M, Wilson, K.S, Wilkinson, A.J, Ab, E, Diercks, T, Folkers, G.E, de Jong, R.N, Truffault, V, Kaptein, R. | | Deposit date: | 2005-07-08 | | Release date: | 2005-09-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of bypass of forespore C, an intercompartmental signaling factor during sporulation in Bacillus.
J. Biol. Chem., 280, 2005
|
|
6VPC
 
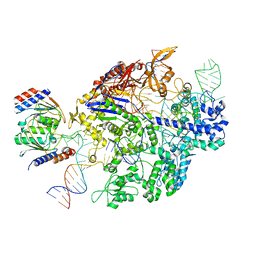 | | Structure of the SpCas9 DNA adenine base editor - ABE8e | | Descriptor: | CRISPR-associated endonuclease Cas9, Cas9 (SpCas9) single-guide RNA (sgRNA), DNA non-target strand (NTS), ... | | Authors: | Knott, G.J, Lapinaite, A, Doudna, J.A. | | Deposit date: | 2020-02-03 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | DNA capture by a CRISPR-Cas9-guided adenine base editor.
Science, 369, 2020
|
|
6VSE
 
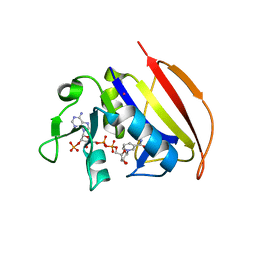 | |
6VV8
 
 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with JEB285 | | Descriptor: | 5-{[3-(1H-indol-3-yl)propanoyl]amino}-1-phenyl-1H-pyrazole-4-carboxylic acid, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Chavez-Pacheco, S.M, Dias, M.V.B. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
1ZOA
 
 | | Crystal Structure Of A328V Mutant Of The Heme Domain Of P450Bm-3 With N-Palmitoylglycine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450:NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Hegda, A, Chen, B, Haines, D.C, Bondlela, M, Mullin, D, Graham, S.E, Tomchick, D.R, Machius, M, Peterson, J.A. | | Deposit date: | 2005-05-12 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A single active-site mutation of P450BM-3 dramatically enhances substrate binding and rate of product formation.
Biochemistry, 50, 2011
|
|
1YXD
 
 | | Structure of E. coli dihydrodipicolinate synthase bound with allosteric inhibitor (S)-lysine to 2.0 A | | Descriptor: | CHLORIDE ION, LYSINE, POTASSIUM ION, ... | | Authors: | Dobson, R.C.J, Griffin, M.D.W, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2005-02-20 | | Release date: | 2005-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structures of native and (S)-lysine-bound dihydrodipicolinate synthase from Escherichia coli with improved resolution show new features of biological significance.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5DFK
 
 | |
1Z2U
 
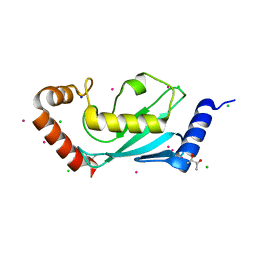 | | The 1.1A crystallographic structure of ubiquitin-conjugating enzyme (ubc-2) from Caenorhabditis elegans: functional and evolutionary significance | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gavira, J.A, DiGiamamarino, E, Tempel, W, Liu, Z.J, Wang, B.C, Meehan, E, Ng, J.D, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-03-09 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1A crystallographic structure of ubiquitin-conjugating enzyme (ubc-2) from Caenorhabditis elegans: functional and evolutionary significance
To be published
|
|
6X4D
 
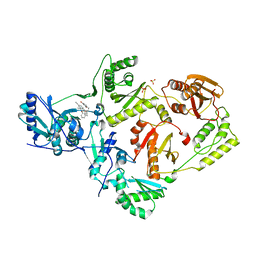 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(cyclopropylmethyl)-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile (JLJ678), a Non-nucleoside Inhibitor | | Descriptor: | 5-(cyclopropylmethyl)-7-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-8-methylnaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
1ZHJ
 
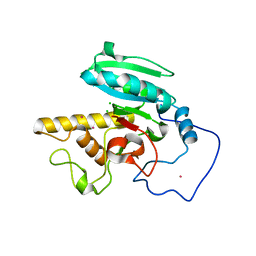 | | Crystal Structure of human N-acetylgalactosaminyltransferase (GTA) Complexed with Galactose | | Descriptor: | CHLORIDE ION, Histo-blood group ABO system transferase (NAGAT) Includes: Glycoprotein-fucosylgalactoside alpha-N- acetylgalactosaminyltransferase, MERCURY (II) ION, ... | | Authors: | Letts, J.A, Rose, N.L, Fang, Y.R, Barry, C.H, Borisova, S.N, Seto, N.O, Palcic, M.M, Evans, S.V. | | Deposit date: | 2005-04-25 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Differential Recognition of the Type I and II H Antigen Acceptors by the Human ABO(H) Blood Group A and B Glycosyltransferases.
J.Biol.Chem., 281, 2006
|
|
5DS5
 
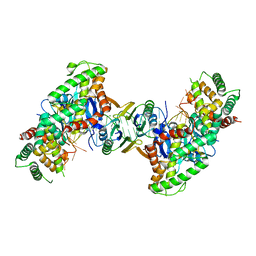 | | Crystal structure the Escherichia coli Cas1-Cas2 complex bound to protospacer DNA and Mg | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | Authors: | Nunez, J.K, Harrington, L.B, Kranzusch, P.J, Engelman, A.N, Doudna, J.A. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Foreign DNA capture during CRISPR-Cas adaptive immunity.
Nature, 527, 2015
|
|
6X1Z
 
 | | Mre11 dimer in complex with small molecule modulator PFMJ | | Descriptor: | (5Z)-5-[(3,4-dimethoxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Arvai, A.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment- and structure-based drug discovery for developing therapeutic agents targeting the DNA Damage Response.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
1ZJF
 
 | | 12mer-spd-P4N | | Descriptor: | 5'-D(*AP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*T)-3', 5'-D(*TP*AP*GP*CP*CP*CP*CP*GP*CP*CP*CP*C)-3' | | Authors: | Dohm, J.A, Hsu, M.H, Hwu, J.R, Huang, R.C, Moudrianakis, E.N, Lattman, E.E, Gittis, A.G. | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Influence of Ions, Hydration, and the Transcriptional Inhibitor P4N on the Conformations of the Sp1 Binding Site.
J.Mol.Biol., 349, 2005
|
|
1ZKH
 
 | | Solution structure of a human ubiquitin-like domain in SF3A1 | | Descriptor: | Splicing factor 3 subunit 1 | | Authors: | Lukin, J.A, Dhe-Paganon, S, Guido, V, Lemak, A, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Sundstrom, M, Edwards, A, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-02 | | Release date: | 2005-05-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a human ubiquitin-like domain in SF3A1
To be Published
|
|
5DMH
 
 | | Crystal structure of a domain of unknown function (DUF1537) from Ralstonia eutropha H16 (H16_A1561), Target EFI-511666, complex with ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Uncharacterized protein conserved in bacteria | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-09-08 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a domain of unknown function (DUF1537) from Ralstonia eutropha H16 (H16_A1561), Target EFI-511666, complex with ADP.
To be published
|
|
6BYR
 
 | | Structures of the PKA RI alpha holoenzyme with the FLHCC driver J-PKAc alpha or native PKAc alpha | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha chimera, MAGNESIUM ION, ... | | Authors: | Cao, B, Lu, T.W, Martinez Fiesco, J.A, Tomasini, M, Fan, L, Simon, S.M, Taylor, S.S, Zhang, P. | | Deposit date: | 2017-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.661 Å) | | Cite: | Structures of the PKA RI alpha Holoenzyme with the FLHCC Driver J-PKAc alpha or Wild-Type PKAc alpha.
Structure, 27, 2019
|
|
5DKE
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a 3-naphthyl-substituted, methyl, cis-diaryl-ethylene compound 4,4'-[2-(naphthalen-2-yl)prop-1-ene-1,1-diyl]diphenol | | Descriptor: | 4,4'-[2-(naphthalen-2-yl)prop-1-ene-1,1-diyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DL4
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a phenylamino-substituted, methyl, triaryl-ethylene derivative 4,4'-{2-[3-(phenylamino)phenyl]prop-1-ene-1,1-diyl}diphenol | | Descriptor: | 4,4'-{2-[3-(phenylamino)phenyl]prop-1-ene-1,1-diyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
6WFL
 
 | | Camphor soaked P450cam D251E | | Descriptor: | 5-EXO-HYDROXYCAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Amaya, J.A, Poulos, T.L, Batabyal, D. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proton Relay Network in the Bacterial P450s: CYP101A1 and CYP101D1.
Biochemistry, 59, 2020
|
|
1ZGV
 
 | | Thrombin in complex with an oxazolopyridine inhibitor 2 | | Descriptor: | Hirudin, N7-BUTYL-N2-(5-CHLORO-2-METHYLPHENYL)-5-METHYL[1,2,4]TRIAZOLO[1,5-A]PYRIMIDINE-2,7-DIAMINE, Thrombin | | Authors: | Deng, J.Z, McMasters, D.R, Rabbat, P.M, Williams, P.D, Coburn, C.A, Yan, Y, Kuo, L.C, Lewis, S.D, Lucas, B.J, Krueger, J.A, Strulovici, B, Vacca, J.P, Lyle, T.A, Burgey, C.S. | | Deposit date: | 2005-04-22 | | Release date: | 2005-09-27 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of an oxazolopyridine series of dual thrombin/factor Xa inhibitors via structure-guided lead optimization.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
5DS4
 
 | | Crystal structure the Escherichia coli Cas1-Cas2 complex bound to protospacer DNA | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER) | | Authors: | Nunez, J.K, Harrington, L.B, Kranzusch, P.J, Engelman, A.N, Doudna, J.A. | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Foreign DNA capture during CRISPR-Cas adaptive immunity.
Nature, 527, 2015
|
|
1ZJ1
 
 | | Crystal Structure of Human Galactosyltransferase (GTB) Complexed with N-acetyllactosamine | | Descriptor: | ABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase), ... | | Authors: | Letts, J.A, Rose, N.L, Fang, Y.R, Barry, C.H, Borisova, S.N, Seto, N.O, Palcic, M.M, Evans, S.V. | | Deposit date: | 2005-04-27 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differential Recognition of the Type I and II H Antigen Acceptors by the Human ABO(H) Blood Group A and B Glycosyltransferases.
J.Biol.Chem., 281, 2006
|
|
1ZJO
 
 | | Crystal Structure of Human N-acetylgalactosaminyltransferase (GTA) Complexed with Galactose-grease | | Descriptor: | CHLORIDE ION, Histo-blood group ABO system transferase (NAGAT) Includes: Glycoprotein-fucosylgalactoside alpha-N- acetylgalactosaminyltransferase, MANGANESE (II) ION, ... | | Authors: | Letts, J.A, Rose, N.L, Fang, Y.R, Barry, C.H, Borisova, S.N, Seto, N.O, Palcic, M.M, Evans, S.V. | | Deposit date: | 2005-04-29 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Differential Recognition of the Type I and II H Antigen Acceptors by the Human ABO(H) Blood Group A and B Glycosyltransferases.
J.Biol.Chem., 281, 2006
|
|
