2Q6J
 
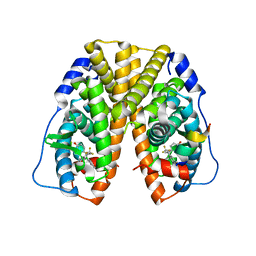 | | Crystal Structure of Estrogen Receptor alpha Complexed to a B-N Substituted Ligand | | Descriptor: | 4-[(DIMESITYLBORYL)(2,2,2-TRIFLUOROETHYL)AMINO]PHENOL, Estrogen receptor, GRIP peptide | | Authors: | Zhou, H, Nettles, K.W, Bruning, J.B, Kim, Y, Joachimiak, A, Sharma, S, Carlson, K.E, Stossi, F, Katzenellenbogen, B.S, Greene, G.L, Katzenellenbogen, J.A. | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Elemental isomerism: a boron-nitrogen surrogate for a carbon-carbon double bond increases the chemical diversity of estrogen receptor ligands
Chem.Biol., 14, 2007
|
|
1B2R
 
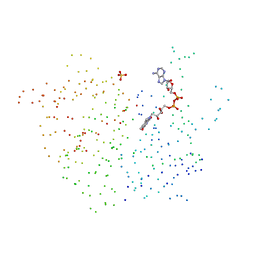 | | FERREDOXIN-NADP+ REDUCTASE (MUTATION: E 301 A) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN-NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-11-27 | | Release date: | 1999-12-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the catalytic role of Glu301 in Anabaena PCC 7119 ferredoxin-NADP+ reductase revealed by x-ray crystallography.
Proteins, 38, 2000
|
|
4C75
 
 | | Consensus (ALL-CON) beta-lactamase class A | | Descriptor: | ACETATE ION, BETA-LACTAMASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2013-09-19 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phenotypic Comparisons of Consensus Variants Versus Laboratory Resurrections of Precambrian Proteins.
Proteins, 82, 2014
|
|
4C94
 
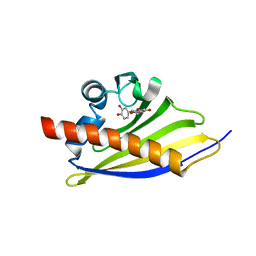 | | Crystal Structure of the Strawberry Pathogenesis-Related 10 (PR-10) Fra a 3 protein in complex with Catechin | | Descriptor: | (2R,3S)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol, FRA A 3 ALLERGEN | | Authors: | Casanal, A, Zander, U, Valpuesta, V, Marquez, J.A. | | Deposit date: | 2013-10-02 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Strawberry Pathogenesis-Related 10 (Pr-10) Fra a Proteins Control Flavonoid Biosynthesis by Binding to Metabolic Intermediates.
J.Biol.Chem., 288, 2013
|
|
2Q87
 
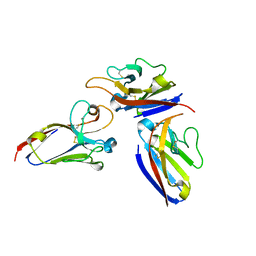 | |
1SSP
 
 | | WILD-TYPE URACIL-DNA GLYCOSYLASE BOUND TO URACIL-CONTAINING DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*(D1P)P*AP*TP*CP*TP*T)-3', URACIL, ... | | Authors: | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
4C6Y
 
 | | Ancestral PNCA (last common ancestors of Gram-positive and Gram- negative bacteria) beta-lactamase class A | | Descriptor: | ACETATE ION, BETA-LACTAMASE, CHLORIDE ION, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M. | | Deposit date: | 2013-09-19 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Phenotypic Comparisons of Consensus Variants Versus Laboratory Resurrections of Precambrian Proteins.
Proteins, 82, 2014
|
|
6B14
 
 | | Crystal structure of Spinach RNA aptamer in complex with Fab BL3-6S97N | | Descriptor: | Heavy chain of Fab BL3-6S97N, Light chain of Fab BL3-6S97N, MAGNESIUM ION, ... | | Authors: | DasGupta, S, Shelke, S.A, Piccirilli, J.A. | | Deposit date: | 2017-09-16 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Affinity maturation of a portable Fab-RNA module for chaperone-assisted RNA crystallography.
Nucleic Acids Res., 46, 2018
|
|
2UWO
 
 | | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa | | Descriptor: | (2R)-2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}PROPENE-1-SULFONAMIDE, COAGULATION FACTOR X | | Authors: | Young, R.J, Brown, D, Burns-Kurtis, C.L, Chan, C, Convery, M.A, Hubbard, J.A, Kelly, H.A, Pateman, A.J, Patikis, A, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Zhou, P. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1AY2
 
 | | STRUCTURE OF THE FIBER-FORMING PROTEIN PILIN AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | HEPTANE-1,2,3-TRIOL, PLATINUM (II) ION, TYPE 4 PILIN, ... | | Authors: | Forest, K.T, Parge, H.E, Tainer, J.A. | | Deposit date: | 1997-11-13 | | Release date: | 1998-04-29 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the fibre-forming protein pilin at 2.6 A resolution.
Nature, 378, 1995
|
|
1T3S
 
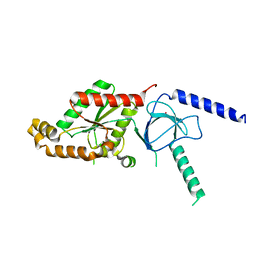 | | Structural Analysis of the Voltage-Dependent Calcium Channel Beta Subunit Functional Core | | Descriptor: | Dihydropyridine-sensitive L-type, calcium channel beta-2 subunit, MERCURY (II) ION | | Authors: | Opatowsky, Y, Chen, C.-C, Campbell, K.P, Hirsch, J.A. | | Deposit date: | 2004-04-27 | | Release date: | 2004-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the voltage-dependent calcium channel beta subunit functional core and its complex with the alpha 1 interaction domain.
Neuron, 42, 2004
|
|
2UWL
 
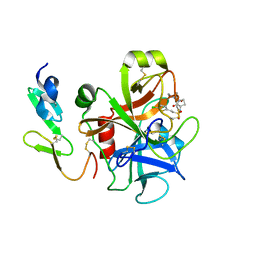 | | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa | | Descriptor: | 2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}ETHENESULFONAMIDE, COAGULATION FACTOR X | | Authors: | Young, R.J, Brown, D, Burns-Kurtis, C.L, Chan, C, Convery, M.A, Hubbard, J.A, Kelly, H.A, Pateman, A.J, Patikis, A, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Zhou, P. | | Deposit date: | 2007-03-22 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6AX6
 
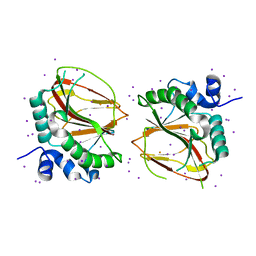 | | The crystal structure of a lysyl hydroxylase from Acanthamoeba polyphaga mimivirus | | Descriptor: | FE (II) ION, IODIDE ION, Procollagen lysyl hydroxylase and glycosyltransferase | | Authors: | Guo, H, Tsai, C, Miller, M.D, Alvarado, S, Tainer, J.A, Phillips Jr, G.N, Kurie, J.M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe2+-binding.
Nat Commun, 9, 2018
|
|
1B38
 
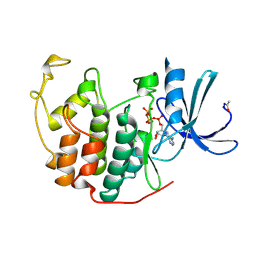 | | HUMAN CYCLIN-DEPENDENT KINASE 2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PROTEIN (CELL DIVISION PROTEIN KINASE 2) | | Authors: | Brown, N.R, Noble, M.E.M, Lawrie, A.M, Morris, M.C, Tunnah, P, Divita, G, Johnson, L.N, Endicott, J.A. | | Deposit date: | 1998-12-17 | | Release date: | 1998-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of phosphorylation of threonine 160 on cyclin-dependent kinase 2 structure and activity.
J.Biol.Chem., 274, 1999
|
|
6AXZ
 
 | |
2V4L
 
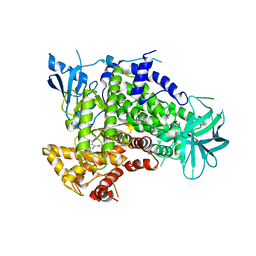 | | complex of human phosphoinositide 3-kinase catalytic subunit gamma (p110 gamma) with PIK-284 | | Descriptor: | 3-[4-AMINO-1-(1-METHYLETHYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL]PHENOL, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Apsel, B, Gonzalez, B, Blair, J.A, Nazif, T.M, Feldman, M.E, Williams, R.L, Shokat, K.M, Knight, Z.A. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeted Polypharmacology: Discovery of Dual Inhibitors of Tyrosine and Phosphoinositide Kinases.
Nat.Chem.Biol., 4, 2008
|
|
2UUK
 
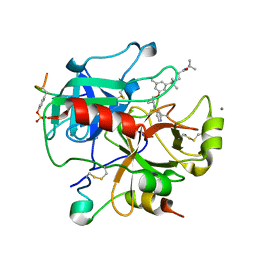 | | Thrombin-hirugen-gw420128 ternary complex at 1.39A resolution | | Descriptor: | CALCIUM ION, HIRUDIN I, HUMAN ALPHA THROMBIN, ... | | Authors: | Ahmed, H.U, Blakeley, M.P, Cianci, M, Cruickshank, D.W.J, Hubbard, J.A, Helliwell, J.R. | | Deposit date: | 2007-03-03 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The Determination of Protonation States in Proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2QAB
 
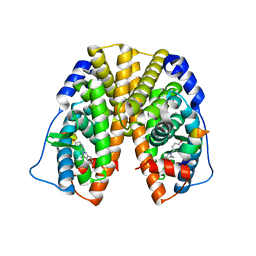 | | Crystal Structure of Estrogen Receptor Alpha Ligand Binding Domain Mutant 537S Complexed with an Ethyl Indazole Compound | | Descriptor: | 3-ETHYL-2-(4-HYDROXYPHENYL)-2H-INDAZOL-5-OL, Estrogen receptor, nuclear receptor coactivator 2 | | Authors: | Nettles, K.W, Bruning, J.B, Nowak, J, Sharma, S.K, Hahm, J.B, Shi, Y, Kulp, K, Hochberg, R.B, Zhou, H, Katzenellenbogen, J.A, Katzenellenbogen, B.S, Kim, Y, Joachmiak, A, Greene, G.L. | | Deposit date: | 2007-06-14 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
2QA6
 
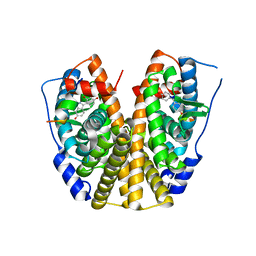 | | Crystal Structure of Estrogen Receptor Alpha mutant 537S Complexed with 4-(6-hydroxy-1H-indazol-3-yl)benzene-1,3-diol | | Descriptor: | 4-(6-HYDROXY-1H-INDAZOL-3-YL)BENZENE-1,3-DIOL, Estrogen receptor, nuclear receptor coactivator 2 | | Authors: | Nettles, K.W, Bruning, J.B, Nowak, J, Sharma, S.K, Hahm, J.B, Shi, Y, Kulp, K, Hochberg, R.B, Zhou, H, Katzenellenbogen, J.A, Katzenellenbogen, B.S, Kim, Y, Joachmiak, A, Greene, G.L. | | Deposit date: | 2007-06-14 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2QS6
 
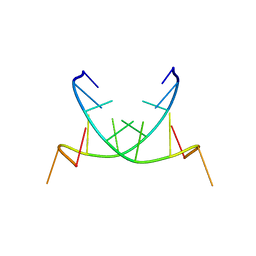 | | Structure of a Hoogsteen antiparallel duplex with extra-helical thymines | | Descriptor: | DNA (5'-D(*DAP*DTP*DAP*DTP*DAP*DTP*DCP*DT)-3') | | Authors: | Pous, J, Urpi, L, Subirana, J.A, Gouyette, C, Navaza, J, Campos, J.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Stabilization by extra-helical thymines of a DNA duplex with Hoogsteen base pairs.
J.Am.Chem.Soc., 130, 2008
|
|
1BYS
 
 | |
2QA7
 
 | |
1BIX
 
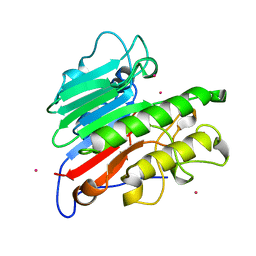 | | THE CRYSTAL STRUCTURE OF THE HUMAN DNA REPAIR ENDONUCLEASE HAP1 SUGGESTS THE RECOGNITION OF EXTRA-HELICAL DEOXYRIBOSE AT DNA ABASIC SITES | | Descriptor: | AP ENDONUCLEASE 1, PLATINUM (II) ION, SAMARIUM (III) ION | | Authors: | Gorman, M.A, Morera, S, Rothwell, D.G, De La Fortelle, E, Mol, C.D, Tainer, J.A, Hickson, I.D, Freemont, P.S. | | Deposit date: | 1998-06-19 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites.
EMBO J., 16, 1997
|
|
2V5U
 
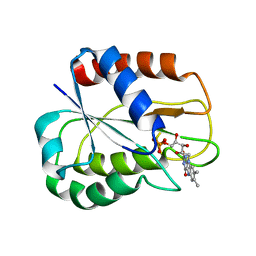 | | I92A FLAVODOXIN FROM ANABAENA | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Martinez-Julvez, M, Herguedas, B, Frago, S, Serrano, A, Molina, R, Hamiaux, C, Schierbeek, B, Medina, M, Hermoso, J.A. | | Deposit date: | 2007-07-10 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Tuning of the Fmn Binding and Oxido-Reduction Properties by Neighboring Side Chains in Anabaena Flavodoxin.
Arch.Biochem.Biophys., 467, 2007
|
|
