1HD4
 
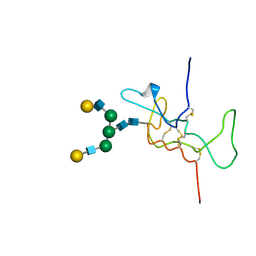 | | SOLUTION STRUCTURE OF THE A-SUBUNIT OF HUMAN CHORIONIC GONADOTROPIN [MODELED WITH DIANTENNARY GLYCAN AT ASN78] | | Descriptor: | CHORIONIC GONADOTROPIN, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Erbel, P.J.A, Karimi-Nejad, Y, van Kuik, J.A, Boelens, R, Kamerling, J.P, Vliegenthart, J.F.G. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-07 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Effects of the N-Linked Glycans on the 3D Structure of the Free A-Subunit of Human Chorionic Gonadotropin
Biochemistry, 39, 2000
|
|
3NN8
 
 | |
4CGN
 
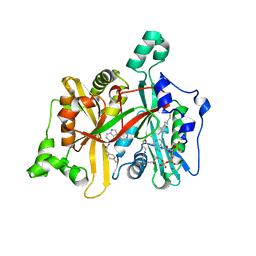 | | Leishmania major N-myristoyltransferase in complex with a piperidinylindole inhibitor | | Descriptor: | 2-(4-fluorophenyl)-N-(3-piperidin-4-yl-1H-indol-5-yl)ethanamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A, Roberts, S.M, Bell, A.S, Hutton, J.A, Smith, D.F, Tate, E.W, Leatherbarrow, R.J, Wilkinson, A.J. | | Deposit date: | 2013-11-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Diverse Modes of Binding in Structures of Leishmania Major N-Myristoyltransferase with Selective Inhibitors
Iucrj, 1, 2014
|
|
4DJB
 
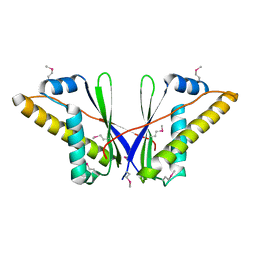 | | A Structural Basis for the Assembly and Functions of a Viral Polymer that Inactivates Multiple Tumor Suppressors | | Descriptor: | E4-ORF3 | | Authors: | Ou, H.D, Kwiatkowski, W, Deerinck, T.J, Noske, A, Blain, K.Y, Land, H.S, Soria, C, Powers, C.J, May, A.P, Shu, X, Tsien, R.Y, Fitzpatrick, J.A.J, Long, J.A, Ellisman, M.H, Choe, S, O'Shea, C.C. | | Deposit date: | 2012-02-01 | | Release date: | 2012-10-31 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | A Structural Basis for the Assembly and Functions of a Viral Polymer that Inactivates Multiple Tumor Suppressors.
Cell(Cambridge,Mass.), 151, 2012
|
|
4M9F
 
 | | Dengue virus NS2B-NS3 protease A125C variant at pH 8.5 | | Descriptor: | NS2B-NS3 protease | | Authors: | Yildiz, M, Ghosh, S, Bell, J.A, Sherman, W, Hardy, J.A. | | Deposit date: | 2013-08-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric Inhibition of the NS2B-NS3 Protease from Dengue Virus.
Acs Chem.Biol., 8, 2013
|
|
4M9T
 
 | |
7PUS
 
 | | ERK5 in complex with Pyrrole Carboxamide scaffold | | Descriptor: | 4-[3,6-bis(chloranyl)-2-fluoranyl-phenyl]carbonyl-~{N}-(1-methylpyrazol-4-yl)-1~{H}-pyrrole-2-carboxamide, Mitogen-activated protein kinase 7 | | Authors: | Tucker, J.A, Martin, M.P, Endicott, J.A, Noble, M.E.N. | | Deposit date: | 2021-09-30 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Parallel Optimization of Potency and Pharmacokinetics Leading to the Discovery of a Pyrrole Carboxamide ERK5 Kinase Domain Inhibitor.
J.Med.Chem., 65, 2022
|
|
6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
8V8L
 
 | | Switchgrass Chalcone Isomerase | | Descriptor: | Chalcone-flavonone isomerase family protein, GLYCEROL | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Interactional Analysis of the Flavonoid Pathway Proteins: Chalcone Synthase, Chalcone Isomerase and Chalcone Isomerase-like Protein.
Int J Mol Sci, 25, 2024
|
|
8V8P
 
 | | Sorghum Chalcone Isomerase | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Chalcone-flavonone isomerase family protein | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structural and Interactional Analysis of the Flavonoid Pathway Proteins: Chalcone Synthase, Chalcone Isomerase and Chalcone Isomerase-like Protein.
Int J Mol Sci, 25, 2024
|
|
8V8N
 
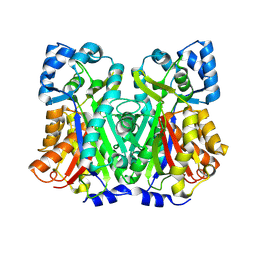 | | Switchgrass Chalcone Synthase C170S | | Descriptor: | Chalcone synthase, GLYCEROL | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and Interactional Analysis of the Flavonoid Pathway Proteins: Chalcone Synthase, Chalcone Isomerase and Chalcone Isomerase-like Protein.
Int J Mol Sci, 25, 2024
|
|
8V8M
 
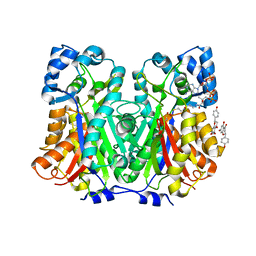 | | Switchgrass Chalcone Synthase | | Descriptor: | COENZYME A, Chalcone synthase, NARINGENIN | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and Interactional Analysis of the Flavonoid Pathway Proteins: Chalcone Synthase, Chalcone Isomerase and Chalcone Isomerase-like Protein.
Int J Mol Sci, 25, 2024
|
|
8V8O
 
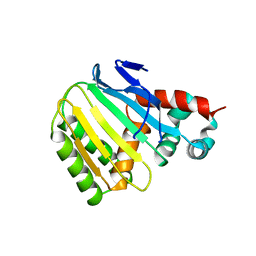 | | Switchgrass Chalcone Isomerase-Like Protein | | Descriptor: | Chalcone-flavonone isomerase family protein | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2023-12-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural and Interactional Analysis of the Flavonoid Pathway Proteins: Chalcone Synthase, Chalcone Isomerase and Chalcone Isomerase-like Protein.
Int J Mol Sci, 25, 2024
|
|
6XKL
 
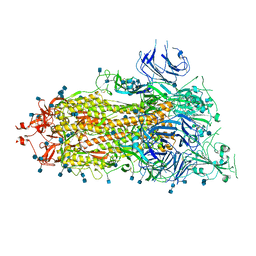 | | SARS-CoV-2 HexaPro S One RBD up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wrapp, D, Hsieh, C.-L, Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure-based design of prefusion-stabilized SARS-CoV-2 spikes.
Science, 369, 2020
|
|
4V6C
 
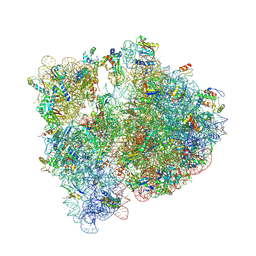 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
4V7V
 
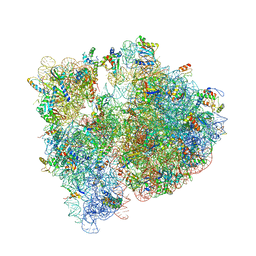 | | Crystal structure of the E. coli ribosome bound to clindamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2891 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V9D
 
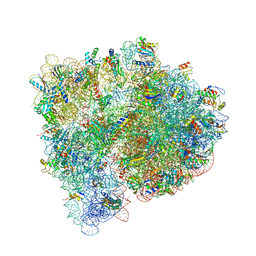 | | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Wang, L, Feldman, M.B, Pulk, A, Chen, V.B, Kapral, G.J, Noeske, J, Richardson, J.S, Blanchard, S.C, Cate, J.H.D. | | Deposit date: | 2012-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding.
Science, 332, 2011
|
|
4UVM
 
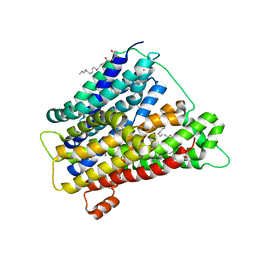 | | In meso crystal structure of the POT family transporter PepTSo | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, GLUTATHIONE UPTAKE TRANSPORTER | | Authors: | Lyons, J.A, Solcan, N, Caffrey, M, Newstead, S. | | Deposit date: | 2014-08-07 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Gating Topology of the Proton-Coupled Oligopeptide Symporters.
Structure, 23, 2015
|
|
5J03
 
 | |
372D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
397D
 
 | |
362D
 
 | | THE STRUCTURE OF D(TGCGCA)2 AND A COMPARISON TO OTHER Z-DNA HEXAMERS | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*TP*GP*CP*GP*CP*A)-3') | | Authors: | Harper, N.A, Brannigan, J.A, Buck, M, Lewis, R.J, Moore, M.H, Schneider, B. | | Deposit date: | 1997-08-20 | | Release date: | 1997-11-20 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of d(TGCGCA)2 and a comparison to other DNA hexamers.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
390D
 
 | | STRUCTURAL VARIABILITY AND NEW INTERMOLECULAR INTERACTIONS OF Z-DNA IN CRYSTALS OF D(PCPGPCPGPCPG) | | Descriptor: | DNA (5'-D(P*CP*GP*CP*GP*CP*G)-3') | | Authors: | Malinina, L, Tereshko, V, Ivanova, E, Subirana, J.A, Zarytova, V, Nekrasov, Y. | | Deposit date: | 1998-04-20 | | Release date: | 1998-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural variability and new intermolecular interactions of Z-DNA in crystals of d(pCpGpCpGpCpG).
Biophys.J., 74, 1998
|
|
5JH2
 
 | | Crystal structure of the holo form of AKR4C7 from maize | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-2'-5'-DIPHOSPHATE, Aldose reductase, ... | | Authors: | Giuseppe, P.O, Santos, M.L, Sousa, S.M, Koch, K.E, Yunes, J.A, Aparicio, R, Murakami, M.T. | | Deposit date: | 2016-04-20 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A comparative structural analysis reveals distinctive features of co-factor binding and substrate specificity in plant aldo-keto reductases.
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5JCG
 
 | |
