3PDS
 
 | | Irreversible Agonist-Beta2 Adrenoceptor Complex | | Descriptor: | 8-hydroxy-5-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(3-sulfanylpropoxy)phenyl]ethyl}amino)ethyl]quinolin-2(1H)-one, CHOLESTEROL, Fusion protein Beta-2 adrenergic receptor/Lysozyme, ... | | Authors: | Rosenbaum, D.M, Zhang, C, Lyons, J.A, Holl, R, Aragao, D, Arlow, D.H, Rasmussen, S.G.F, Choi, H.-J, DeVree, B.T, Sunahara, R.K, Chae, P.S, Gellman, S.H, Dror, R.O, Shaw, D.E, Weis, W.I, Caffrey, M, Gmeiner, P, Kobilka, B.K. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and function of an irreversible agonist-beta(2) adrenoceptor complex
Nature, 469, 2011
|
|
3PBW
 
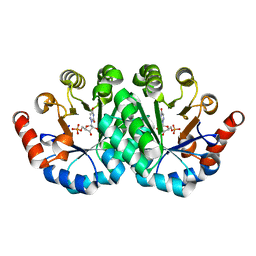 | | Crystal structure of the mutant L123N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
3PBU
 
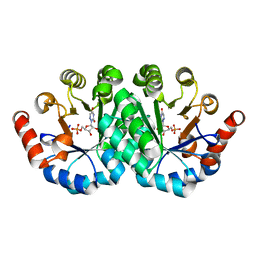 | | Crystal structure of the mutant I96S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
3PC0
 
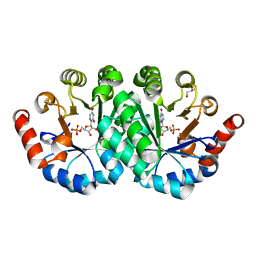 | | Crystal structure of the mutant V155S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
4JQZ
 
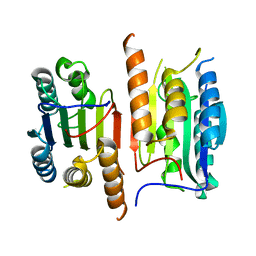 | | Human procaspase-3, crystal form 2 | | Descriptor: | Procaspase-3 | | Authors: | Thomsen, N.D, Wells, J.A. | | Deposit date: | 2013-03-20 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | Structural snapshots reveal distinct mechanisms of procaspase-3 and -7 activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4J9X
 
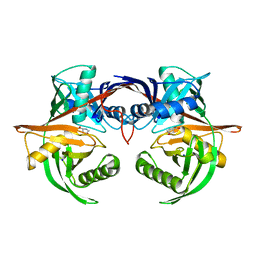 | | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with trans-4-hydroxy-l-proline | | Descriptor: | 4-HYDROXYPROLINE, Proline racemase family protein, SODIUM ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-17 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with trans-4-hydroxy-l-proline
To be Published
|
|
1S45
 
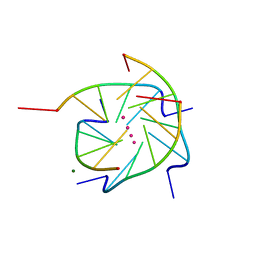 | | Crystal structure analysis of the DNA quadruplex d(TGGGGT) S1 | | Descriptor: | 5'-D(*TP*GP*GP*GP*GP*T)-3', MAGNESIUM ION, SODIUM ION, ... | | Authors: | Caceres, C, Wright, G, Gouyette, C, Parkinson, G, Subirana, J.A. | | Deposit date: | 2004-01-15 | | Release date: | 2004-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Thymine tetrad in d(TGGGGT) quadruplexes stabilized with Tl+1/Na+1 ions
Nucleic Acids Res., 32, 2004
|
|
4JDP
 
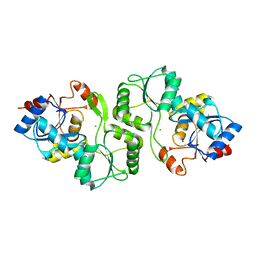 | | Crystal structure of probable p-nitrophenyl phosphatase (pho2) (target EFI-501307) from Archaeoglobus fulgidus DSM 4304 with Magnesium bound | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, p-nitrophenyl phosphatase (Pho2) | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Allen, K.N, Dunaway-Mariano, D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of probable p-nitrophenyl phosphatase from Archaeoglobus fulgidus.
To be Published
|
|
1S5W
 
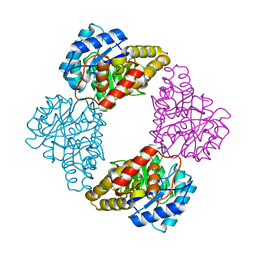 | |
1S69
 
 | | The X-ray structure of the cyanobacteria Synechocystis hemoglobin "cyanoglobin" with cyanide ligand | | Descriptor: | CITRATE ANION, CYANIDE ION, Cyanoglobin, ... | | Authors: | Trent III, J.T, Kundu, S, Hoy, J.A, Hargrove, M.S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystallographic analysis of synechocystis cyanoglobin reveals the structural changes accompanying ligand binding in a hexacoordinate hemoglobin.
J.Mol.Biol., 341, 2004
|
|
4JFX
 
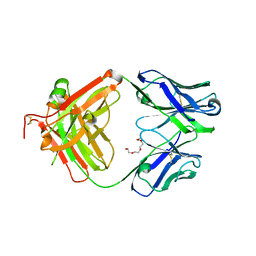 | | Structure of phosphotyrosine (pTyr) scaffold bound to pTyr peptide | | Descriptor: | Fab heavy chain, Fab light chain, Phosphopeptide, ... | | Authors: | Koerber, J.T, Thomsen, N.D, Hannigan, B.T, Degrado, W.F, Wells, J.A. | | Deposit date: | 2013-02-28 | | Release date: | 2013-09-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Nature-inspired design of motif-specific antibody scaffolds.
Nat.Biotechnol., 31, 2013
|
|
1SCE
 
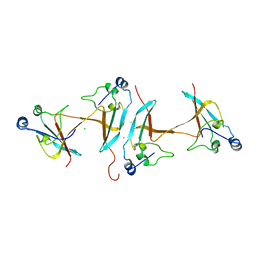 | |
4JPX
 
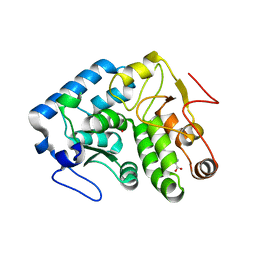 | |
1SKK
 
 | | Structure of the antimicrobial hexapeptide cyc-(KKWWKF) bound to DPC micelles | | Descriptor: | cyclic hexapeptide KKWWKF | | Authors: | Appelt, C, Soderhall, J.A, Bienert, M, Dathe, M, Schmieder, P. | | Deposit date: | 2004-03-05 | | Release date: | 2005-03-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the antimicrobial, cationic hexapeptide cyclo(RRWWRF) and its analogues in solution and bound to detergent micelles.
Chembiochem, 6, 2005
|
|
1SH0
 
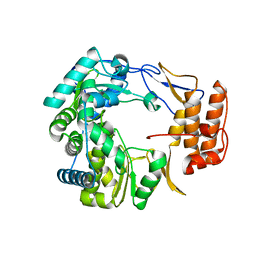 | | Crystal Structure of Norwalk Virus Polymerase (Triclinic) | | Descriptor: | RNA Polymerase | | Authors: | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
1P6L
 
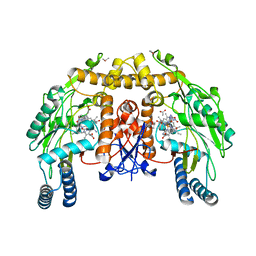 | | Bovine endothelial NOS heme domain with L-N(omega)-nitroarginine-2,4-L-diaminobutyric amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1OTC
 
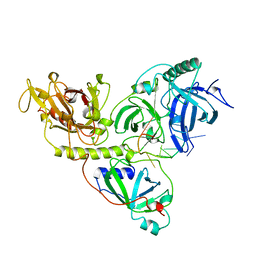 | | THE O. NOVA TELOMERE END BINDING PROTEIN COMPLEXED WITH SINGLE STRAND DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3'), PROTEIN (TELOMERE-BINDING PROTEIN ALPHA SUBUNIT), PROTEIN (TELOMERE-BINDING PROTEIN BETA SUBUNIT) | | Authors: | Horvath, M.P, Schweiker, V.L, Bevilacqua, J.M, Ruggles, J.A, Schultz, S.C. | | Deposit date: | 1998-11-25 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA.
Cell(Cambridge,Mass.), 95, 1998
|
|
3PSG
 
 | |
1OXQ
 
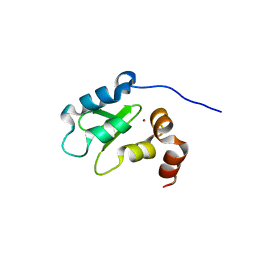 | | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, AVPIAQKSE (Smac) peptide, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Kadkhodayan, S, Ackerly, H, Alexandru, D, Distefano, M.D, Elliott, L.O, Flygare, J.A, Vucic, D, Deshayes, K, Fairbrother, W.J. | | Deposit date: | 2003-04-03 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP)
Biochemistry, 42, 2003
|
|
3Q72
 
 | | Crystal Structure of Rad G-domain-GTP Analog Complex | | Descriptor: | CALCIUM ION, GTP-binding protein RAD, MAGNESIUM ION, ... | | Authors: | Sasson, Y, Navon-Perry, L, Hirsch, J.A. | | Deposit date: | 2011-01-04 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | RGK Family G-Domain:GTP Analog Complex Structures and Nucleotide-Binding Properties.
J.Mol.Biol., 413, 2011
|
|
3Q7P
 
 | |
1OZT
 
 | | Crystal Structure of apo-H46R Familial ALS Mutant human Cu,Zn Superoxide Dismutase (CuZnSOD) to 2.5A resolution | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Elam, J.S, Taylor, A.B, Strange, R, Antonyuk, S, Doucette, P.A, Rodriguez, J.A, Hasnain, S.S, Hayward, L.J, Valentine, J.S, Yeates, T.O, Hart, P.J. | | Deposit date: | 2003-04-09 | | Release date: | 2003-05-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Amyloid-like Filaments and Water-filled Nanotubes Formed by SOD1 Mutant Proteins Linked to Familial ALS
Nat.Struct.Biol., 10, 2003
|
|
1P6H
 
 | | Rat neuronal NOS heme domain with L-N(omega)-nitroarginine-2,4-L-diaminobutyric amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, L-N(OMEGA)-NITROARGININE-2,4-L-DIAMINOBUTYRIC AMIDE, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
4KDX
 
 | | Crystal structure of a glutathione transferase family member from burkholderia graminis, target efi-507264, bound gsh, ordered domains, space group p21, form(1) | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase domain | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-25 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a glutathione transferase family member from burkholderia graminis, target efi-507264, bound gsh, ordered domains, space group P21, form(1)
To be Published
|
|
3R09
 
 | | Crystal structure of probable HAD family hydrolase from Pseudomonas fluorescens Pf-5 with bound Mg | | Descriptor: | Hydrolase, haloacid dehalogenase-like family, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-03-07 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of probable HAD family hydrolase from Pseudomonas fluorescens Pf-5 with bound Mg
To be Published
|
|
