3NMQ
 
 | | Hsp90b N-terminal domain in complex with EC44, a pyrrolo-pyrimidine methoxypyridine inhibitor | | Descriptor: | 5-{2-amino-4-chloro-7-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-7H-pyrrolo[2,3-d]pyrimidin-5-yl}-2-methylpent-4-yn-2 -ol, Heat shock protein HSP 90-beta | | Authors: | Arndt, J.W, Yun, T.J, Harning, E.K, Giza, K, Rabah, D, Li, P, Luchetti, D, Shi, J, Manning, A, Kehry, M.R. | | Deposit date: | 2010-06-22 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | EC144, a Synthetic Inhibitor of Heat Shock Protein 90, Blocks Innate and Adaptive Immune Responses in Models of Inflammation and Autoimmunity.
J.Immunol., 186, 2011
|
|
3NN1
 
 | | Structure of chlorite dismutase from Candidatus Nitrospira defluvii in complex with imidazole | | Descriptor: | 1,2-ETHANEDIOL, Chlorite dismutase, IMIDAZOLE, ... | | Authors: | Kostan, J, Sjoeblom, B, Maixner, F, Mlynek, G, Furtmueller, P.G, Obinger, C, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": Identification of a catalytically important amino acid residue
J.Struct.Biol., 172, 2010
|
|
3NA9
 
 | | Crystal structure of Fab15 | | Descriptor: | ACETATE ION, Fab15 heavy chain, Fab15 light chain, ... | | Authors: | Luo, J. | | Deposit date: | 2010-06-01 | | Release date: | 2010-08-18 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-evolution of antibody stability and Vk CDR-L3 canonical structure
To be Published
|
|
3N8E
 
 | | Substrate binding domain of the human Heat Shock 70kDa protein 9 (mortalin) | | Descriptor: | Stress-70 protein, mitochondrial | | Authors: | Wisniewska, M, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, van der Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate binding domain of the human Heat Shock 70kDa protein 9 (mortalin)
To be published
|
|
3NFC
 
 | |
3NHV
 
 | | Crystal Structure of BH2092 protein from Bacillus halodurans, Northeast Structural Genomics Consortium Target BhR228F | | Descriptor: | BH2092 protein, CHLORIDE ION | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-11 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Northeast Structural Genomics Consortium Target BhR228F
To be Published, 2010
|
|
3NML
 
 | | Sperm whale myoglobin mutant H64W carbonmonoxy-form | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Birukou, I, Soman, J, Olson, J.S. | | Deposit date: | 2010-06-22 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Blocking the gate to ligand entry in human hemoglobin.
J.Biol.Chem., 286, 2011
|
|
3NDK
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dCTP Opposite dG | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-06-07 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Variation in Mutation Rates Caused by RB69pol Fidelity Mutants Can Be Rationalized on the Basis of Their Kinetic Behavior and Crystal Structures.
J.Mol.Biol., 406, 2011
|
|
3NMV
 
 | | Crystal structure of pyrabactin-bound abscisic acid receptor PYL2 mutant A93F in complex with type 2C protein phosphatase ABI2 | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NN8
 
 | |
3NF7
 
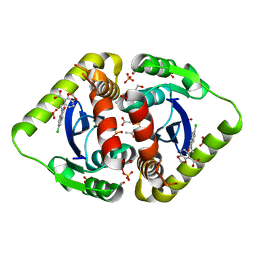 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | Descriptor: | 1,2-ETHANEDIOL, 5-[(5-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3N9A
 
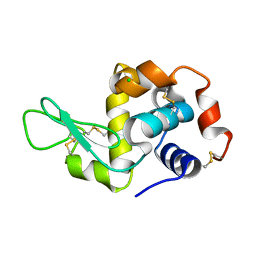 | |
3N9C
 
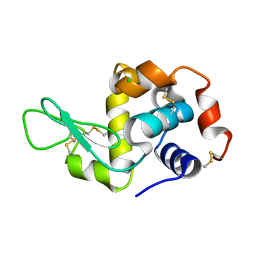 | | Mite-y Lysozyme: Marmite | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Newman, J, Peat, T.S. | | Deposit date: | 2010-05-28 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mite-y Lysozyme Crystals and Structures
To be Published
|
|
3NAA
 
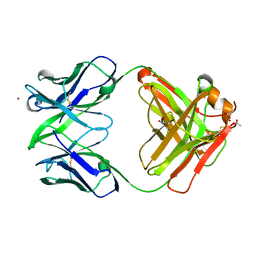 | | Crystal structure of Fab15 Mut5 | | Descriptor: | ACETATE ION, Fab15 Mut5 heavy chain, Fab15 Mut5 light chain, ... | | Authors: | Luo, J, Feng, Y, Gilliland, G.L. | | Deposit date: | 2010-06-01 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-evolution of antibody stability and Vk CDR-L3 canonical structure
To be Published
|
|
3NMM
 
 | | Human Hemoglobin A mutant alpha H58W deoxy-form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Birukou, I, Soman, J, Olson, J.S. | | Deposit date: | 2010-06-22 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Blocking the gate to ligand entry in human hemoglobin.
J.Biol.Chem., 286, 2011
|
|
3NF8
 
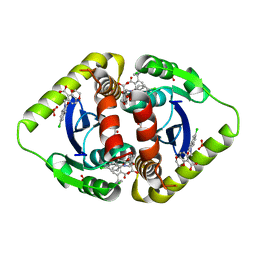 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | Descriptor: | 6-[(5-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETIC ACID, Integrase, ... | | Authors: | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3NFK
 
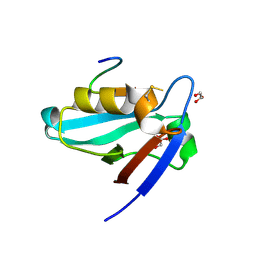 | | Crystal structure of the PTPN4 PDZ domain complexed with the C-terminus of a rabies virus G protein | | Descriptor: | GLYCEROL, Glycoprotein G, Tyrosine-protein phosphatase non-receptor type 4 | | Authors: | Babault, N, Cordier, F, Lafage, M, Cockburn, J, Haouz, A, Rey, F.A, Delepierre, M, Buc, H, Lafon, M, Wolff, N. | | Deposit date: | 2010-06-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Peptides Targeting the PDZ Domain of PTPN4 Are Efficient Inducers of Glioblastoma Cell Death.
Structure, 19, 2011
|
|
3NEH
 
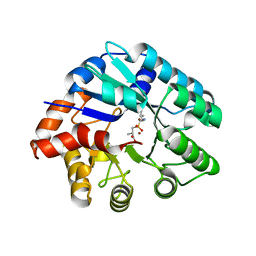 | | Crystal structure of the protein LMO2462 from Listeria monocytogenes complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala | | Descriptor: | (2R)-3-[(R)-[(1R)-1-amino-3-methylbutyl](hydroxy)phosphoryl]-2-methylpropanoic acid, Renal dipeptidase family protein, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-08 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Crystal structure of the protein LMO2462 from Listeria monocytogenes
complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala
To be Published
|
|
3NFA
 
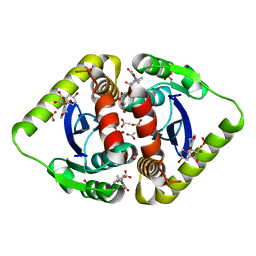 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | Descriptor: | 6-[(5-bromo-2,3-dioxo-2,3-dihydro-1H-indol-1-yl)methyl]-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETIC ACID, Integrase, ... | | Authors: | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | Deposit date: | 2010-06-10 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3NFL
 
 | | Crystal structure of the PTPN4 PDZ domain complexed with the C-terminus of the GluN2A NMDA receptor subunit | | Descriptor: | Glutamate [NMDA] receptor subunit epsilon-1, Tyrosine-protein phosphatase non-receptor type 4 | | Authors: | Babault, N, Cordier, F, Lafage, M, Cockburn, J, Haouz, A, Rey, F.A, Delepierre, M, Buc, H, Lafon, M, Wolff, N. | | Deposit date: | 2010-06-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Peptides Targeting the PDZ Domain of PTPN4 Are Efficient Inducers of Glioblastoma Cell Death.
Structure, 19, 2011
|
|
3NKG
 
 | | Crystal Structure of GEBA250068378 from Sulfurospirillum deleyianum | | Descriptor: | ACETIC ACID, GLYCEROL, uncharacterized protein GEBA250068378 | | Authors: | Kim, Y, Marshall, N, Cobb, G, Eisen, J, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-19 | | Release date: | 2010-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of GEBA250068378 from Sulfurospirillum deleyianum
To be Published, 2010
|
|
3NHX
 
 | | Crystal Structure of Ketosteroid Isomerase D99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Ruben, E, Sunden, F, Herschlag, D. | | Deposit date: | 2010-06-14 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound
To be Published
|
|
3NN3
 
 | | Structure of chlorite dismutase from Candidatus Nitrospira defluvii R173A mutant | | Descriptor: | Chlorite dismutase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kostan, J, Sjoeblom, B, Maixner, F, Mlynek, G, Furtmueller, P.G, Obinger, C, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium "Candidatus Nitrospira defluvii": Identification of a catalytically important amino acid residue
J.Struct.Biol., 172, 2010
|
|
3NIX
 
 | | Crystal structure of flavoprotein/dehydrogenase from Cytophaga hutchinsonii. Northeast Structural Genomics Consortium Target ChR43. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavoprotein/dehydrogenase | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Foote, E.L, Ciccosanti, C, Maglaqui, M, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of flavoprotein/dehydrogenase from Cytophaga hutchinsonii.
To be Published
|
|
3NMN
 
 | | Crystal structure of pyrabactin-bound abscisic acid receptor PYL1 in complex with type 2C protein phosphatase ABI1 | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL1, MAGNESIUM ION, ... | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
