1YOK
 
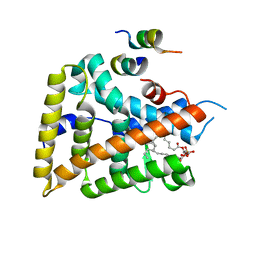 | | crystal structure of human LRH-1 bound with TIF-2 peptide and phosphatidylglycerol | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Nuclear receptor coactivator 2, Orphan nuclear receptor NR5A2 | | Authors: | Krylova, I.N, Sablin, E.P, Moore, J, Xu, R.X, Waitt, G.M, MacKay, J.A, Juzumiene, D, Bynum, J.M, Madauss, K, Montana, V, Lebedeva, L, Suzawa, M, Williams, J.D, Williams, S.P, Guy, R.K, Thornton, J.W, Fletterick, R.J, Willson, T.M, Ingraham, H.A. | | Deposit date: | 2005-01-27 | | Release date: | 2005-07-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1.
Cell(Cambridge,Mass.), 120, 2005
|
|
1YOX
 
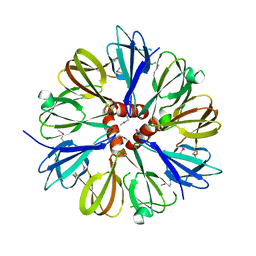 | | Structure of the conserved Protein of Unknown Function PA3696 from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein PA3696 | | Authors: | Walker, J.R, Xu, X, Gu, J, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-26 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of the conserved hypothetical protein PA3696
To be Published
|
|
1YRL
 
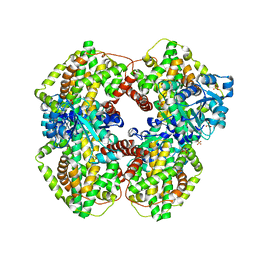 | | Escherichia coli ketol-acid reductoisomerase | | Descriptor: | Ketol-acid reductoisomerase, SULFATE ION | | Authors: | Tyagi, R, Duquerroy, S, Navaza, J, Guddat, L.W, Duggleby, R.G. | | Deposit date: | 2005-02-04 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of a bacterial class II ketol-acid reductoisomerase: domain conservation and evolution
Protein Sci., 14, 2005
|
|
1YSM
 
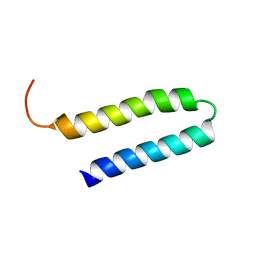 | | NMR Structure of N-terminal domain (Residues 1-77) of Siah-Interacting Protein. | | Descriptor: | Calcyclin-binding protein | | Authors: | Bhattacharya, S, Lee, Y.T, Michowski, W, Jastrzebska, B, Filipek, A, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2005-02-08 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Modular Structure of SIP Facilitates Its Role in Stabilizing Multiprotein Assemblies.
Biochemistry, 44, 2005
|
|
5DHP
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a novel inhibitor | | Descriptor: | 8-({2-oxo-2-[(2-phenylethyl)amino]ethyl}sulfanyl)adenosine, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Paoletti, J, Assairi, L, Huteau, V, Pochet, S, Labesse, G. | | Deposit date: | 2015-08-31 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | 8-Thioalkyl-adenosine derivatives inhibit Listeria monocytogenes NAD kinase through a novel binding mode.
Eur.J.Med.Chem., 124, 2016
|
|
4RHN
 
 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT COMPLEXED WITH ADENOSINE | | Descriptor: | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN, alpha-D-ribofuranose | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-26 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
1YOU
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with a potent pyrimidinetrione inhibitor | | Descriptor: | 5-(2-ETHOXYETHYL)-5-[4-(4-FLUOROPHENOXY)PHENOXY]PYRIMIDINE-2,4,6(1H,3H,5H)-TRIONE, CALCIUM ION, Collagenase 3, ... | | Authors: | Pandit, J. | | Deposit date: | 2005-01-28 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent pyrimidinetrione-based inhibitors of MMP-13 with enhanced selectivity over MMP-14.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1YP9
 
 | | Trypsin Inhibitor Complex | | Descriptor: | (1R,3AS,4R,8AS,8BR)-4-(2-BENZO[1,3]DIOXOL-5-YLMETHYL)-1-BENZYL-3-OXO-DECAHYDRO-PYRROLO[3,4-A]PYRROZILIN-4-YL-BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Fokkens, J, Obst-Sander, U, Heine, A, Diederich, F, Klebe, G. | | Deposit date: | 2005-01-31 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A simple protocol to estimate differences in protein binding affinity for enantiomers without prior resolution of racemates
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
4RHP
 
 | | Crystal structure of human COQ9 in complex with a phospholipid, Northeast Structural Genomics Consortium Target HR5043 | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Ubiquinone biosynthesis protein COQ9, mitochondrial | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Wang, H, Lee, D, Kogan, S, Maglaqui, M, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-22 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1YPJ
 
 | | Thrombin Inhibitor Complex | | Descriptor: | (1R,3AS,4R,8AS,8BR)-4-{5-(PHENYL[1,3]DIOXOL-5-YLMETHYL)-4-ETHYL-2,3,3-TRIMETHYL-6-OXO-OCTAHYDRO-PYRROLO[3,4-C]PYRROL-1-YL}-BENZAMIDINE, Hirudin, Thrombin heavy chain, ... | | Authors: | Fokkens, J, Obst-Sander, U, Heine, A, Diederich, F, Klebe, G. | | Deposit date: | 2005-01-31 | | Release date: | 2006-01-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A simple protocol to estimate differences in protein binding affinity for enantiomers without prior resolution of racemates
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
8IRL
 
 | | Apo state of Arabidopsis AZG1 at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRN
 
 | | 6-BAP bound state of Arabidopsis AZG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1, N-BENZYL-9H-PURIN-6-AMINE | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRP
 
 | | kinetin bound state of Arabidopsis AZG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1, N-(FURAN-2-YLMETHYL)-7H-PURIN-6-AMINE | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8IRM
 
 | | Endogenous substrate adenine bound state of Arabidopsis AZG1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
4RJV
 
 | | Crystal Structure of a De Novo Designed Ferredoxin Fold, Northeast Structural Genomics Consortium (NESG) Target OR461 | | Descriptor: | OR461 | | Authors: | O'Connell, P.T, Lin, Y.-R, Guan, R, Koga, N, Koga, R, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-10-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR461
To be published
|
|
1YRX
 
 | | Structure of a novel photoreceptor: the BLUF domain of AppA from Rhodobacter sphaeroides | | Descriptor: | FLAVIN MONONUCLEOTIDE, N-DODECYL-N,N-DIMETHYLGLYCINATE, hypothetical protein Rsph03001874 | | Authors: | Anderson, S, Dragnea, V, Masuda, S, Ybe, J, Moffat, K, Bauer, C. | | Deposit date: | 2005-02-05 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Novel Photoreceptor, the BLUF Domain of AppA from Rhodobacter sphaeroides
Biochemistry, 44, 2005
|
|
8IRO
 
 | | trans-Zeatin bound state of Arabidopsis AZG1 at pH7.4 | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8J22
 
 | | Cryo-EM structure of FFAR2 complex bound with TUG-1375 | | Descriptor: | (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]carbonyl-1,3-thiazolidine-4-carboxylic acid, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J20
 
 | | Cryo-EM structure of FFAR3 bound with valeric acid and AR420626 | | Descriptor: | (4R)-N-[2,5-bis(chloranyl)phenyl]-4-(furan-2-yl)-2-methyl-5-oxidanylidene-4,6,7,8-tetrahydro-1H-quinoline-3-carboxamide, Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
1A5E
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 18 STRUCTURES | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
8J24
 
 | | Cryo-EM structure of FFAR2 complex bound with acetic acid | | Descriptor: | ACETATE ION, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Tang, W, Sun, X, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
1YPM
 
 | |
4RKM
 
 | | Wolinella succinogenes octaheme sulfite reductase MccA, form I | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ACETATE ION, COPPER (I) ION, ... | | Authors: | Hermann, B, Kern, M, La Pietra, L, Simon, J, Einsle, O. | | Deposit date: | 2014-10-13 | | Release date: | 2015-02-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The octahaem MccA is a haem c-copper sulfite reductase.
Nature, 520, 2015
|
|
1YRH
 
 | |
1YU7
 
 | | Crystal Structure of the W64Y mutant of Villin Headpiece | | Descriptor: | Villin | | Authors: | Meng, J, Vardar, D, Wang, Y, Guo, H.C, Head, J.F, McKnight, C.J. | | Deposit date: | 2005-02-12 | | Release date: | 2005-09-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structures of villin headpiece and mutants with reduced F-actin binding activity.
Biochemistry, 44, 2005
|
|
