8PH1
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3708 | | Descriptor: | 7-[(1~{S})-1-[(5~{R})-5-[(2~{R})-3-azanyl-2-methyl-propyl]-2-oxidanylidene-1,3-oxazolidin-3-yl]ethyl]-3-(1~{H}-pyrazol-4-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3708
To Be Published
|
|
8PGI
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2984 | | Descriptor: | 7-[(1~{S})-1-[5-[(3-azanyl-3-methyl-azetidin-1-yl)methyl]-2-oxidanylidene-1,3-oxazolidin-3-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2984
To Be Published
|
|
8PH0
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3637 | | Descriptor: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-(2-oxidanylidene-1,3-dihydroindol-5-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3637
To Be Published
|
|
6V2S
 
 | | Crystal Structure of chromodomain of MPP8 in complex with inhibitor UNC3866 | | Descriptor: | M-phase phosphoprotein 8, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
8PGJ
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2985 | | Descriptor: | 7-[(1~{S})-1-[5-[(3-azanylazetidin-1-yl)methyl]-2-oxidanylidene-1,3-oxazolidin-3-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, DIMETHYL SULFOXIDE, ... | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2985
To Be Published
|
|
8PG5
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2731 | | Descriptor: | 3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-7-[(1~{S})-1-(piperidin-4-ylmethoxy)ethyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2731
To Be Published
|
|
7T1T
 
 | | JAK2 JH2 IN COMPLEX WITH JAK292 | | Descriptor: | (2S)-2-[({4-[(2-amino-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy]phenyl}carbamoyl)amino]-4-phenylbutanoic acid, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Ippolito, J.A, Henry, S, Krimmer, S.G, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2021-12-02 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Conversion of a False Virtual Screen Hit into Selective JAK2 JH2 Domain Binders Using Convergent Design Strategies
Acs Med.Chem.Lett., 13, 2022
|
|
2K37
 
 | | CsmA | | Descriptor: | Chlorosome Protein A | | Authors: | Pedersen, M, Dittmer, J, Underhaug, J, Miller, M, Nielsen, N.C. | | Deposit date: | 2008-04-23 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of CsmA: A small antenna protein from the green sulfur bacterium Chlorobium tepidum.
Febs Lett., 582, 2008
|
|
8PG7
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2755 | | Descriptor: | 7-[(1~{S})-1-[3-(4-azanylbutyl)-2,5-bis(oxidanylidene)imidazolidin-1-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2755
To Be Published
|
|
8PGW
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3460 | | Descriptor: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-(5-methoxy-6-oxidanyl-pyridin-3-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3460
To Be Published
|
|
8PJD
 
 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 94-445 | | Descriptor: | ACETATE ION, Protein arginine N-methyltransferase 2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2023-06-23 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 94-445
To Be Published
|
|
8PM7
 
 | |
7E3S
 
 | |
8PMC
 
 | |
5UG3
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN GID MUTANT A10V | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A.K, Leffler, A.E, Zebroski, H.A, Powell, S.R, Kuryatov, A, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UG5
 
 | | NMR SOLUTION STRUCTURE OF THE ALPHA-CONOTOXIN GID MUTANT V13Y | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A, Leffler, A.E, Kuryatov, A, Zebroski, H.A, Powell, S.R, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8PUV
 
 | | Trimeric prM/E spike of Tick-borne encephalitis virus immature particle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Protein prM | | Authors: | Fuzik, T, Plevka, P, Smerdova, L, Novacek, J. | | Deposit date: | 2023-07-17 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The structure of immature tick-borne encephalitis virus supports the collapse model of flavivirus maturation.
Sci Adv, 10, 2024
|
|
7E3R
 
 | | Crystal structure of TrmL from Vibrio vulnificus | | Descriptor: | tRNA (cytidine(34)-2'-O)-methyltransferase | | Authors: | Kim, J, Son, J. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of tRNA mehtyltransferase TrmL from Vibrio vulnificus
To be published
|
|
6V4P
 
 | | Structure of the integrin AlphaIIbBeta3-Abciximab complex | | Descriptor: | Abciximab, heavy chain, light chain, ... | | Authors: | Nesic, D, Zhang, Y, Spasic, A, Li, J, Provasi, D, Filizola, M, Walz, T, Coller, B.S. | | Deposit date: | 2019-11-28 | | Release date: | 2020-02-05 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-Electron Microscopy Structure of the alpha IIb beta 3-Abciximab Complex.
Arterioscler Thromb Vasc Biol., 40, 2020
|
|
7E3T
 
 | |
6UVY
 
 | | BACE-1 in complex with compound #18 | | Descriptor: | (1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-N-{[(1R,2R)-2-methylcyclopropyl]methyl}cyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
7AVK
 
 | | Streptococcal High Identity Repeats in Tandem (SHIRT) domain 10 from cell surface protein SGO_0707 | | Descriptor: | LPXTG cell wall surface protein, isothiocyanate | | Authors: | Degut, C, Gilburt, J, Whelan, F, Jenkins, H.T, Potts, J.R. | | Deposit date: | 2020-11-05 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (0.82 Å) | | Cite: | Periscope Proteins are variable-length regulators of bacterial cell surface interactions.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SR8
 
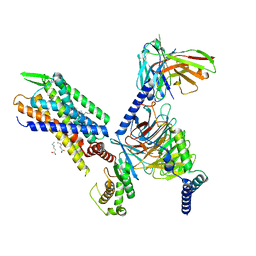 | | Molecular mechanism of the the wake-promoting agent TAK-925 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Hypocretin receptor type 2, ... | | Authors: | Yin, J, Chapman, K, Lian, P, De Brabander, J.K, Rosenbaum, D.M. | | Deposit date: | 2021-11-08 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanism of the wake-promoting agent TAK-925.
Nat Commun, 13, 2022
|
|
3O0A
 
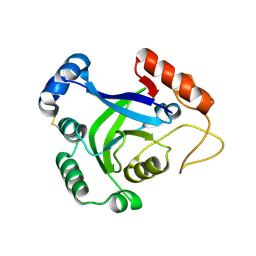 | | Crystal structure of the wild type CP1 hydrolitic domain from Aquifex Aeolicus leucyl-trna | | Descriptor: | Leucyl-tRNA synthetase subunit alpha | | Authors: | Cura, V, Olieric, N, Wang, E.-D, Moras, D, Eriani, G, Cavarelli, J. | | Deposit date: | 2010-07-19 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of the Wild Type and Two Mutants of the Cp1 Hydrolytic Domain from Aquifex Aeolicus Leucyl-tRNA Synthetase
To be Published
|
|
6UZ1
 
 | |
