4UQK
 
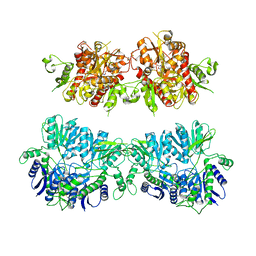 | | Electron density map of GluA2em in complex with quisqualate and LY451646 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2 | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4URD
 
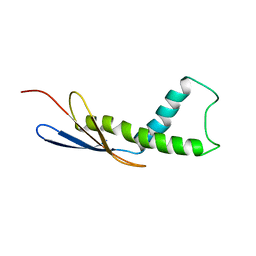 | | Cryo-EM map of Trigger Factor bound to a translating ribosome | | Descriptor: | TRIGGER FACTOR | | Authors: | Deeng, J, Chan, K.Y, van der Sluis, E, Bischoff, L, Berninghausen, O, Han, W, Gumbart, J, Schulten, K, Beatrix, B, Beckmann, R. | | Deposit date: | 2014-06-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Dynamic Behavior of Trigger Factor on the Ribosome.
J.Mol.Biol., 428, 2016
|
|
6YUR
 
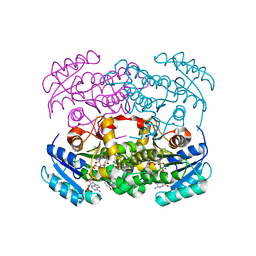 | | Crystal structure of S. aureus FabI inhibited by SKTS1 | | Descriptor: | 6-[4-(4-hexyl-2-oxidanyl-phenoxy)phenoxy]pyridin-2-ol, Enoyl-[acyl-carrier-protein] reductase [NADPH], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Weinrich, J.D, Eltschkner, S, Schiebel, J, Kehrein, J, Le, T.A, Davoodi, S, Merget, B, Tonge, P.J, Engels, B, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A Long Residence Time Enoyl-Reductase Inhibitor Explores an Extended Binding Region with Isoenzyme-Dependent Tautomer Adaptation and Differential Substrate-Binding Loop Closure.
Acs Infect Dis., 7, 2021
|
|
5HI8
 
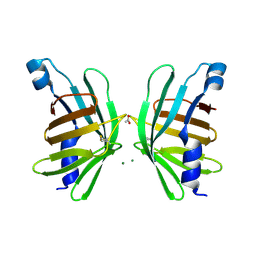 | | Structure of T-type Phycobiliprotein Lyase CpeT from Prochlorococcus phage P-HM1 | | Descriptor: | ACETATE ION, Antenna protein, MAGNESIUM ION | | Authors: | Gasper, R, Schwach, J, Frankenberg-Dinkel, N, Hofmann, E. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct Features of Cyanophage-encoded T-type Phycobiliprotein Lyase Phi CpeT: THE ROLE OF AUXILIARY METABOLIC GENES.
J. Biol. Chem., 292, 2017
|
|
6YUU
 
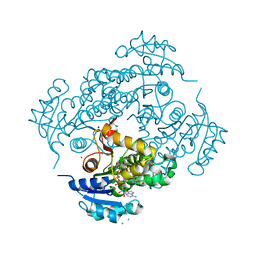 | | Crystal structure of M. tuberculosis InhA inhibited by SKTS1 | | Descriptor: | 6-[4-(4-hexyl-2-oxidanyl-phenoxy)phenoxy]pyridin-2-ol, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Schiebel, J, Kehrein, J, Le, T.A, Davoodi, S, Merget, B, Weinrich, J.D, Tonge, P.J, Engels, B, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A Long Residence Time Enoyl-Reductase Inhibitor Explores an Extended Binding Region with Isoenzyme-Dependent Tautomer Adaptation and Differential Substrate-Binding Loop Closure.
Acs Infect Dis., 7, 2021
|
|
4V6M
 
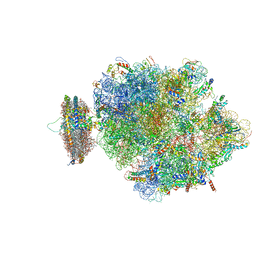 | | Structure of the ribosome-SecYE complex in the membrane environment | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 16S RIBOSOMAL RNA, ... | | Authors: | Frauenfeld, J, Gumbart, J, van der Sluis, E.O, Funes, S, Gartmann, M, Beatrix, B, Mielke, T, Berninghausen, O, Becker, T, Schulten, K, Beckmann, R. | | Deposit date: | 2011-02-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Cryo-EM structure of the ribosome-SecYE complex in the membrane environment.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4UZC
 
 | |
3T0H
 
 | |
3T0Z
 
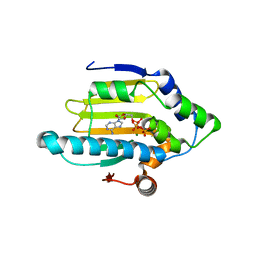 | | Hsp90 N-terminal domain bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Li, J. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90.
Acta Biochim Biophys Sin (Shanghai), 44, 2012
|
|
2EIS
 
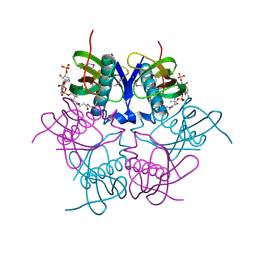 | | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8 | | Descriptor: | COENZYME A, Hypothetical protein TTHB207 | | Authors: | Kamitori, S, Yoshida, H, Satoh, S, Iino, H, Ebihara, A, Chen, L, Fu, Z.-Q, Chrzas, J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-13 | | Release date: | 2008-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8
To be Published
|
|
2DYF
 
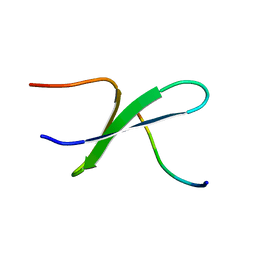 | |
4V96
 
 | | The structure of a 1.8 MDa viral genome injection device suggests alternative infection mechanisms | | Descriptor: | BPP, ORF46, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V, van Sinderen, D, Cambillau, C. | | Deposit date: | 2012-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4Z5P
 
 | | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.9 A resolution | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, TRIETHYLENE GLYCOL | | Authors: | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-02 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
4EGX
 
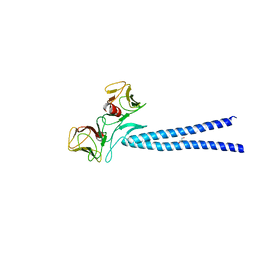 | | Crystal structure of KIF1A CC1-FHA tandem | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Kinesin-like protein KIF1A | | Authors: | Yu, J, Huo, L, Yue, Y, Xu, T, Zhang, M, Feng, W. | | Deposit date: | 2012-04-02 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The CC1-FHA Tandem as a Central Hub for Controlling the Dimerization and Activation of Kinesin-3 KIF1A
Structure, 20, 2012
|
|
2DXS
 
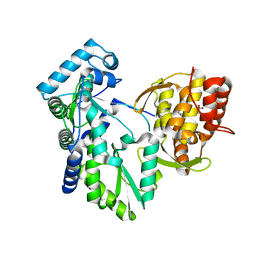 | | Crystal structure of HCV NS5B RNA polymerase complexed with a tetracyclic inhibitor | | Descriptor: | Genome polyprotein, N-[(13-CYCLOHEXYL-6,7-DIHYDROINDOLO[1,2-D][1,4]BENZOXAZEPIN-10-YL)CARBONYL]-2-METHYL-L-ALANINE | | Authors: | Adachi, T, Tsuruha, J, Doi, S, Murase, K, Ikegashira, K, Watanabe, S, Uehara, K, Orita, T, Nomura, A, Kamada, M. | | Deposit date: | 2006-08-30 | | Release date: | 2006-12-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Conformationally Constrained Tetracyclic Compounds as Potent Hepatitis C Virus NS5B RNA Polymerase Inhibitors
J.Med.Chem., 49, 2006
|
|
4EQC
 
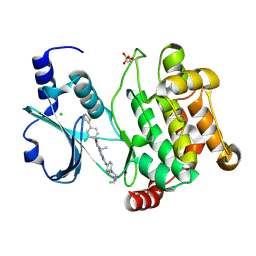 | | Crystal structure of PAK1 kinase domain in complex with FRAX597 inhibitor | | Descriptor: | 6-[2-chloro-4-(1,3-thiazol-5-yl)phenyl]-8-ethyl-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, CHLORIDE ION, Serine/threonine-protein kinase PAK 1 | | Authors: | Maksimoska, J, Marmorstein, R. | | Deposit date: | 2012-04-18 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | FRAX597, a Small Molecule Inhibitor of the p21-activated Kinases, Inhibits Tumorigenesis of Neurofibromatosis Type 2 (NF2)-associated Schwannomas.
J.Biol.Chem., 288, 2013
|
|
4V6S
 
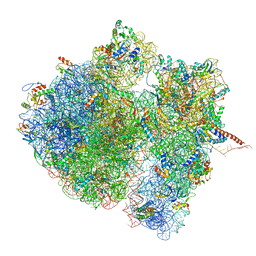 | | Structural characterization of mRNA-tRNA translocation intermediates (class 3 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (13.1 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V82
 
 | | Crystal structure of cyanobacterial Photosystem II in complex with terbutryn | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Gabdulkhakov, A, Broser, M, Guskov, A, Kern, J, Glockner, C, Muh, F, Saenger, W, Zouni, A. | | Deposit date: | 2010-11-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of cyanobacterial photosystem II Inhibition by the herbicide terbutryn
J.Biol.Chem., 286, 2011
|
|
2EC8
 
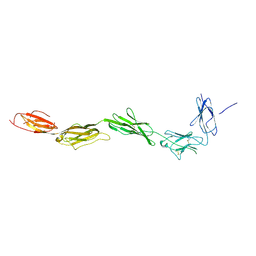 | | Crystal structure of the exctracellular domain of the receptor tyrosine kinase, Kit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Mast/stem cell growth factor receptor | | Authors: | Yuzawa, S, Opatowsky, Y, Zhang, Z, Mandiyan, V, Lax, I, Schlessinger, J. | | Deposit date: | 2007-02-11 | | Release date: | 2007-08-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Activation of the Receptor Tyrosine Kinase KIT by Stem Cell Factor
Cell(Cambridge,Mass.), 130, 2007
|
|
6Z71
 
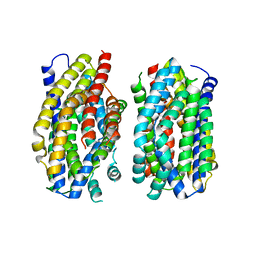 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Z70
 
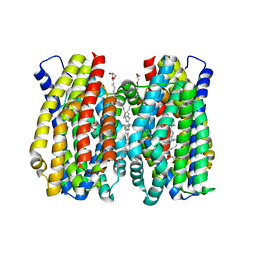 | | Structure of the MATE family multidrug resistance transporter Aq_128 from Aquifex aeolicus in the outward-facing state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Aq128 | | Authors: | Zhao, J, Safarian, S, Thielmann, Y, Xie, H, Wang, J, Michel, H. | | Deposit date: | 2020-05-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the Aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4WCZ
 
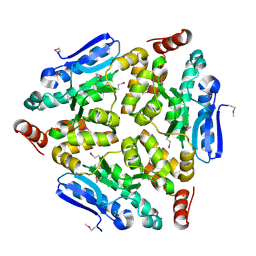 | | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Chapman, H.C, Niedzialkowska, E, Cymborowski, M.T, Hillerich, B.S, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans
to be published
|
|
2EBI
 
 | | Arabidopsis GT-1 DNA-binding domain with T133D phosphomimetic mutation | | Descriptor: | DNA binding protein GT-1 | | Authors: | Nagata, T, Noto, K, Niyada, E, Ikeda, Y, Yamamoto, Y, Uesugi, S, Murata, J, Hiratsuka, K, Katahira, M. | | Deposit date: | 2007-02-08 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: mechanism for an increase in DNA-binding affinity through phosphorylation.
Proteins, 78, 2010
|
|
4V6O
 
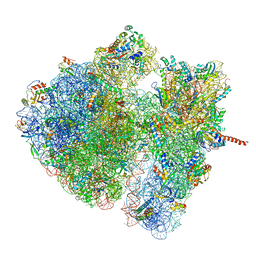 | | Structural characterization of mRNA-tRNA translocation intermediates (class 4a of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2E26
 
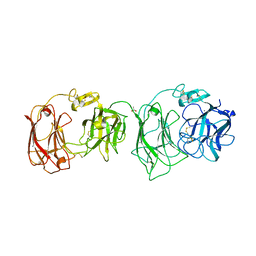 | | Crystal structure of two repeat fragment of reelin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Yasui, N, Nogi, T, Kitao, T, Takagi, J. | | Deposit date: | 2006-11-08 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a receptor-binding fragment of reelin and mutational analysis reveal a recognition mechanism similar to endocytic receptors.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
