2PID
 
 | | Crystal structure of human mitochondrial tyrosyl-tRNA synthetase in complex with an adenylate analog | | Descriptor: | 5'-O-[N-(L-TYROSYL)SULFAMOYL]ADENOSINE, Tyrosyl-tRNA synthetase | | Authors: | Bonnefond, L, Frugier, M, Touze, E, Lorber, B, Florentz, C, Giege, R, Sauter, C, Rudinger-Thirion, J. | | Deposit date: | 2007-04-13 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Human Mitochondrial Tyrosyl-tRNA Synthetase Reveals Common and Idiosyncratic Features.
Structure, 15, 2007
|
|
6K4M
 
 | |
6K60
 
 | | Structural and functional basis for HLA-G isoform recognition of immune checkpoint receptor LILRBs | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain G, ... | | Authors: | Kuroki, K, Matsubara, H, Kanda, R, Miyashita, N, Shiroishi, M, Fukunaga, Y, Kamishikiryo, J, Fukunaga, A, Hirose, K, Sugita, Y, Kita, S, Ose, T, Maenaka, K. | | Deposit date: | 2019-05-31 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.149 Å) | | Cite: | Structural and Functional Basis for LILRB Immune Checkpoint Receptor Recognition of HLA-G Isoforms.
J Immunol., 203, 2019
|
|
6K40
 
 | | Crystal structure of alkyl hydroperoxide reductase from D. radiodurans R1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alkyl hydroperoxide reductase AhpD, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, M.-K, Zhang, J, Zhao, L. | | Deposit date: | 2019-05-22 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of the AhpD-like protein DR1765 from Deinococcus radiodurans R1.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
2VVI
 
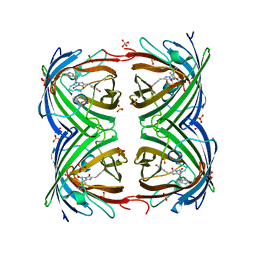 | | IrisFP fluorescent protein in its green form, trans conformation | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION, SULFITE ION | | Authors: | Adam, V, Lelimousin, M, Boehme, S, Desfonds, G, Nienhaus, K, Field, M.J, Wiedenmann, J, McSweeney, S, Nienhaus, G.U, Bourgeois, D. | | Deposit date: | 2008-06-09 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Irisfp, an Optical Highlighter Undergoing Multiple Photo-Induced Transformations.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5OJF
 
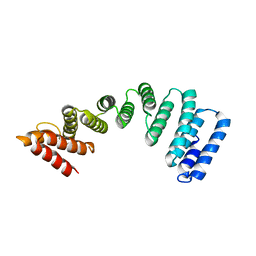 | | Crystal Structure of KLC2-TPR domain (fragment [A1-B6] | | Descriptor: | Kinesin light chain 2 | | Authors: | Nguyen, T.Q, Chenon, M, Vilela, F, Velours, C, Andreani, J, Fernandez-Varela, P, Llinas, P, Menetrey, J. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural plasticity of the N-terminal capping helix of the TPR domain of kinesin light chain.
PLoS ONE, 12, 2017
|
|
6K4X
 
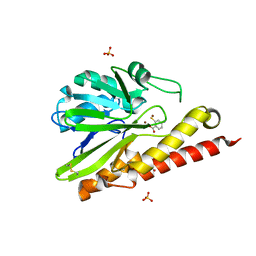 | | Crystal structure of SMB-1 metallo-beta-lactamase in a complex with ASB | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-azanyl-2-sulfanyl-benzoic acid, Metallo-beta-lactamase, ... | | Authors: | Wachino, J. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | 4-Amino-2-Sulfanylbenzoic Acid as a Potent Subclass B3 Metallo-beta-Lactamase-Specific Inhibitor Applicable for Distinguishing Metallo-beta-Lactamase Subclasses.
Antimicrob.Agents Chemother., 63, 2019
|
|
2VYC
 
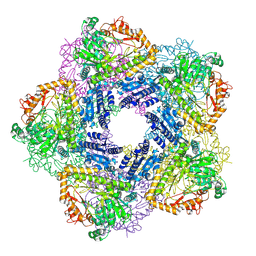 | | Crystal Structure of Acid Induced Arginine Decarboxylase from E. coli | | Descriptor: | BIODEGRADATIVE ARGININE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Andrell, J, Hicks, M.G, Palmer, T, Carpenter, E.P, Iwata, S, Maher, M.J. | | Deposit date: | 2008-07-22 | | Release date: | 2009-03-31 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Acid Induced Arginine Decarboxylase from Escherichia Coli: Reversible Decamer Assembly Controls Enzyme Activity.
Biochemistry, 48, 2009
|
|
6K57
 
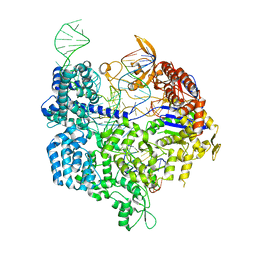 | | Crystal structure of dCas9 in complex with sgRNA and DNA (CGA PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9, non-target DNA, sgRNA, ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-28 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
2VVU
 
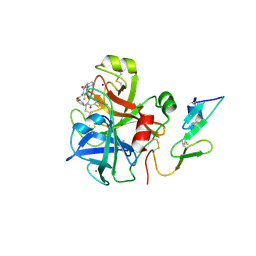 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-chloro-N-[(3R)-1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)pyrrolidin-3-yl]thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-11 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2W75
 
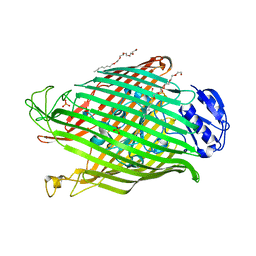 | | Structures of P. aeruginosa FpvA bound to heterologous pyoverdines: Apo-FpvA | | Descriptor: | 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, FERRIPYOVERDINE RECEPTOR, PHOSPHATE ION | | Authors: | Greenwald, J, Nader, M, Celia, H, Gruffaz, C, Meyer, J.-M, Schalk, I.J, Pattus, F. | | Deposit date: | 2008-12-20 | | Release date: | 2009-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Fpva Bound to Non-Cognate Pyoverdines: Molecular Basis of Siderophore Recognition by an Iron Transporter.
Mol.Microbiol., 72, 2009
|
|
2WB6
 
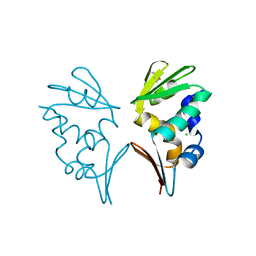 | | Crystal structure of AFV1-102, a protein from the Acidianus Filamentous Virus 1 | | Descriptor: | AFV1-102, CHLORIDE ION | | Authors: | Keller, J, Leulliot, N, Collinet, B, Campanacci, V, Cambillau, C, Pranghisvilli, D, van Tilbeurgh, H. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Afv1-102, a Protein from the Acidianus Filamentous Virus 1.
Protein Sci., 18, 2009
|
|
7FH0
 
 | |
2WV7
 
 | | Intracellular subtilisin precursor from B. clausii | | Descriptor: | INTRACELLULAR SUBTILISIN PROTEASE, SODIUM ION | | Authors: | Vevodova, J, Gamble, M, Ariza, A, Dodson, E, Jones, D.D, Wilson, K.S. | | Deposit date: | 2009-10-15 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of an Intracellular Subtilisin Reveals Novel Structural Features Unique to This Subtilisin Family.
Structure, 18, 2010
|
|
2WTR
 
 | |
2WWH
 
 | | Plasmodium falciparum thymidylate kinase in complex with AP5dT | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, SODIUM ION, THYMIDILATE KINASE, ... | | Authors: | Whittingham, J.L, Carrero-Lerida, J, Brannigan, J.A, Ruiz-Perez, L.M, Silva, A.P.G, Fogg, M.J, Wilkinson, A.J, Gilbert, I.H, Wilson, K.S, Gonzalez-Pacanowska, D. | | Deposit date: | 2009-10-23 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Efficient Phosphorylation of Aztmp and Dgmp by Plasmodium Falciparum Type I Thymidylate Kinase.
Biochem.J., 428, 2010
|
|
2WV4
 
 | | Crystal structure of foot-and-mouth disease virus 3C protease in complex with a decameric peptide corresponding to the VP1-2A cleavage junction | | Descriptor: | FOOT AND MOUTH DISEASE VIRUS (SEROTYPE A) VARIANT VP1 CAPSID PROTEIN, PICORNAIN 3C | | Authors: | Zunszain, P.A, Knox, S.R, Sweeney, T.R, Yang, J, Roque-Rosell, N, Belsham, G.J, Leatherbarrow, R.J, Curry, S. | | Deposit date: | 2009-10-13 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into Cleavage Specificity from the Crystal Structure of Foot-and-Mouth Disease Virus 3C Protease Complexed with a Peptide Substrate.
J.Mol.Biol., 395, 2010
|
|
4EKX
 
 | |
6KI8
 
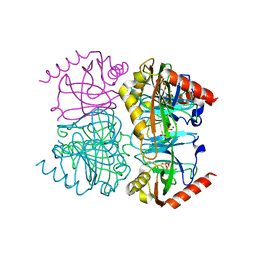 | | Pyrophosphatase mutant K149R from Acinetobacter baumannii | | Descriptor: | DIPHOSPHATE, Inorganic pyrophosphatase, MAGNESIUM ION | | Authors: | Su, J. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structures of Pyrophosphatase from Acinetobacter baumannii: Snapshots of Pyrophosphate Binding and Identification of a Phosphorylated Enzyme Intermediate.
Int J Mol Sci, 20, 2019
|
|
6KF3
 
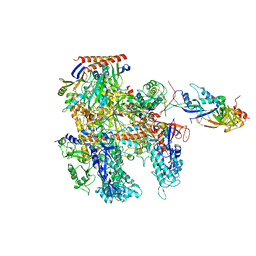 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
6KIZ
 
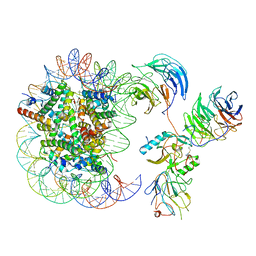 | | Cryo-EM structure of human MLL1-NCP complex, binding mode2 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
2Q06
 
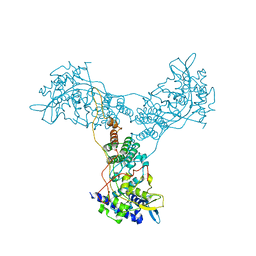 | | Crystal structure of Influenza A Virus H5N1 Nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Ng, A.K.L, Zhang, H, Tan, K, Wang, J, Shaw, P.C. | | Deposit date: | 2007-05-21 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design.
Faseb J., 22, 2008
|
|
2X7G
 
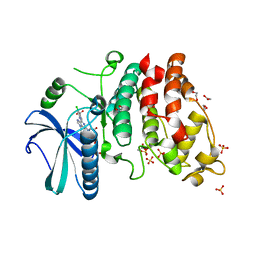 | | Structure of human serine-arginine-rich protein-specific kinase 2 (SRPK2) bound to purvalanol B | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PURVALANOL B, ... | | Authors: | Pike, A.C.W, Savitsky, P, Fedorov, O, Krojer, T, Ugochukwu, E, von Delft, F, Gileadi, O, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Human Serine-Arginine-Rich Protein- Specific Kinase 2 (Srpk2) Bound to Purvalanol B
To be Published
|
|
6KN1
 
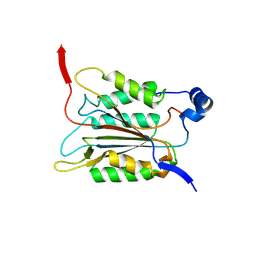 | | P20/P12 of caspase-11 mutant C254A | | Descriptor: | Caspase-4 | | Authors: | Ding, J, Sun, Q. | | Deposit date: | 2019-08-02 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis.
Cell, 180, 2020
|
|
6KIU
 
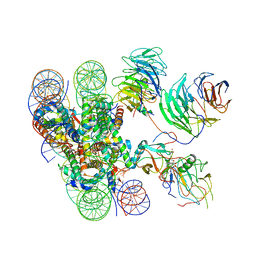 | | Cryo-EM structure of human MLL1-ubNCP complex (3.2 angstrom) | | Descriptor: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
