8E41
 
 | | E. coli 50S ribosome bound to tiamulin and VS1 | | Descriptor: | 50S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Tiamulin and NPET binders
To Be Published
|
|
8E3M
 
 | | E. coli 50S ribosome bound to L-linker solithromycin conjugate | | Descriptor: | 50S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Solithromycin siderophore conjugates
To Be Published
|
|
8E44
 
 | | E. coli 50S ribosome bound to antibiotic analog SLC09 | | Descriptor: | 2-chloro-N-{[(5S)-3-{(4M)-3-fluoro-4-[(6P)-6-(2-methyl-2H-tetrazol-5-yl)pyridin-3-yl]phenyl}-2-oxo-1,3-oxazolidin-5-yl]methyl}acetamide, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | SLC collection of antibiotic analogs
To Be Published
|
|
4Z4P
 
 | | Structure of the MLL4 SET Domain | | Descriptor: | Histone-lysine N-methyltransferase 2D, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Zhang, Z, Mittal, A, Reid, J, Reich, S, Gamblin, S.J, Wilson, J.R. | | Deposit date: | 2015-04-02 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolving Catalytic Properties of the MLL Family SET Domain.
Structure, 23, 2015
|
|
4BTA
 
 | | CRYSTAL STRUCTURE OF THE PEPTIDE(PRO-PRO-GLY)3 BOUND COMPLEX OF N- TERMINAL DOMAIN AND PEPTIDE SUBSTRATE BINDING DOMAIN OF PROLYL-4 HYDROXYLASE (RESIDUES 1-244) TYPE I FROM HUMAN | | Descriptor: | PROLINE RICH PEPTIDE, PROLYL 4-HYDROXYLASE SUBUNIT ALPHA-1 | | Authors: | Anantharajan, J, Koski, M.K, Pekkala, M, Wierenga, R.K. | | Deposit date: | 2013-06-14 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The Structural Motifs for Substrate Binding and Dimerization of the Alpha Subunit of Collagen Prolyl 4-Hydroxylase
Structure, 21, 2013
|
|
4Z51
 
 | | High Resolution Human Septin 3 GTPase domain | | Descriptor: | MAGNESIUM ION, Neuronal-specific septin-3, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Valadares, N.F, Macedo, J.N, Leonardo, D.A, Brandao-Neto, J, Pereira, H.M, Matos, S.O, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2015-04-02 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A complete compendium of crystal structures for the human SEPT3 subgroup reveals functional plasticity at a specific septin interface
Iucrj, 2020
|
|
8E42
 
 | | E. coli 50S ribosome bound to tiamulin and azithromycin | | Descriptor: | 50S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Tiamulin and NPET binders
To Be Published
|
|
8E35
 
 | | E. coli 50S ribosome bound to compound SAB002 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-16 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Potential for clicked streptogramin analogs
To Be Published
|
|
8E47
 
 | | E. coli 50S ribosome bound to antibiotic analog SLC26 | | Descriptor: | (2R,3S,4R,5R,8R,10R,11R,12S,13S,14R)-2-ethyl-3,4,10-trihydroxy-3,5,6,8,10,12,14-heptamethyl-15-oxo-11-({3,4,6-trideoxy-3-[(2-{[2-({[(5S)-3-{(4M)-3-fluoro-4-[(6P)-6-(2-methyl-2H-tetrazol-5-yl)pyridin-3-yl]phenyl}-2-oxo-1,3-oxazolidin-5-yl]methyl}amino)-2-oxoethyl]sulfanyl}ethyl)(methyl)amino]-beta-D-xylo-hexopyranosyl}oxy)-1-oxa-6-azacyclopentadecan-13-yl 2,6-dideoxy-3-C-methyl-3-O-methyl-alpha-L-ribo-hexopyranoside, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | SLC collection of antibiotic analogs
To Be Published
|
|
8E3O
 
 | | E. coli 50S ribosome bound to solithromycin and VM1 | | Descriptor: | (3aS,4R,7S,9R,10R,11R,13R,15R,15aR)-1-{4-[4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl]butyl}-4-ethyl-7-fluoro-11-methoxy-3a ,7,9,11,13,15-hexamethyl-2,6,8,14-tetraoxotetradecahydro-2H-oxacyclotetradecino[4,3-d][1,3]oxazol-10-yl 3,4,6-trideoxy-3-(dimethylamino)-beta-D-xylo-hexopyranoside, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (1.99 Å) | | Cite: | Solithromycin siderophore conjugates
To Be Published
|
|
4BT8
 
 | |
4Z9L
 
 | | THE STRUCTURE OF JNK3 IN COMPLEX WITH AN IMIDAZOLE-PYRIMIDINE INHIBITOR | | Descriptor: | CYCLOHEXYL-{4-[5-(3,4-DICHLOROPHENYL)-2-PIPERIDIN-4-YL-3-PROPYL-3H-IMIDAZOL-4-YL]-PYRIMIDIN-2-YL}AMINE, Mitogen-activated protein kinase 10, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, Lograsso, P.V, Smart, O.S, Bricogne, G. | | Deposit date: | 2015-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity.
Chem.Biol., 10, 2003
|
|
4ZCK
 
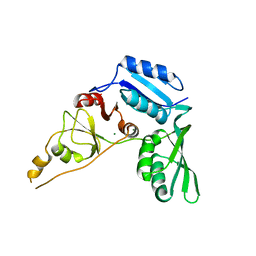 | | Crystal Structure of C-terminal Fragment of Escherichia coli BipA/TypA | | Descriptor: | GTP-binding protein TypA/BipA, MAGNESIUM ION | | Authors: | Fan, H.T, Hahm, J, Diggs, S, Blaha, G. | | Deposit date: | 2015-04-16 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural and Functional Analysis of BipA, a Regulator of Virulence in Enteropathogenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
4ZCM
 
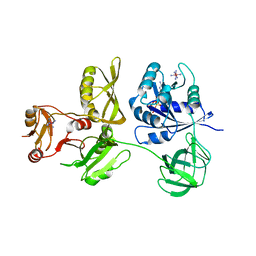 | | Crystal Structure of Escherichia coli GTPase BipA/TypA Complexed with ppGpp | | Descriptor: | COBALT HEXAMMINE(III), GTP-binding protein TypA/BipA, GUANOSINE-5',3'-TETRAPHOSPHATE, ... | | Authors: | Fan, H.T, Hahm, J, Diggs, S, Blaha, G. | | Deposit date: | 2015-04-16 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural and Functional Analysis of BipA, a Regulator of Virulence in Enteropathogenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
8CWP
 
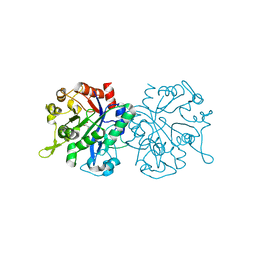 | | X-ray crystal structure of NTHi Protein D bound to a putative glycerol moiety | | Descriptor: | GLYCEROL, Glycerophosphoryl diester phosphodiesterase, SODIUM ION | | Authors: | Jones, S.P, Cook, K.H, Holmquist, M.L, Almekinder, L, DeLaney, A, Labbe, N, Perdue, J, Jackson, N, Charles, R, Pichichero, M, Kaur, R, Michel, L, Gleghorn, M.L. | | Deposit date: | 2022-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Vaccine target and carrier molecule nontypeable Haemophilus influenzae protein D dimerizes like the close Escherichia coli GlpQ homolog but unlike other known homolog dimers.
Proteins, 91, 2023
|
|
4Z0B
 
 | | Crystal Structure of the Fab Fragment of Anti-ofloxacin Antibody and Exploration Its Receptor Binding Site | | Descriptor: | PHOSPHATE ION, antibody heavy chain, antibody light chain | | Authors: | He, K, Du, X, Sheng, W, Zhou, X, Wang, J, Wang, S. | | Deposit date: | 2015-03-26 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Fab Fragment of an Anti-ofloxacin Antibody and Exploration of Its Specific Binding.
J.Agric.Food Chem., 64, 2016
|
|
4Z4Q
 
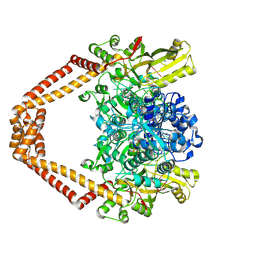 | | Quinazolinedione(PD 0305970)-DNA cleavage complex of topoisomerase IV from S. pneumoniae | | Descriptor: | 3-amino-7-{(3R)-3-[(1S)-1-aminoethyl]pyrrolidin-1-yl}-1-cyclopropyl-6-fluoro-8-methylquinazoline-2,4(1H,3H)-dione, DNA topoisomerase 4 subunit B,DNA topoisomerase 4 subunit A, MAGNESIUM ION, ... | | Authors: | Laponogov, I, Veselkov, D.A, Pan, X.-S, Selvarajah, J, Crevel, I.M.-T, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2015-04-02 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural studies of the drug-stabilized cleavage complexes of topoisomerase IV and gyrase from Streptococcus pneumoniae
To Be Published
|
|
4I07
 
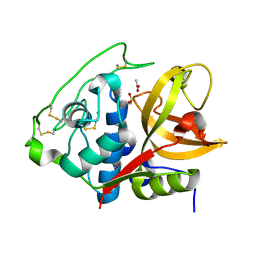 | | Structure of mature form of cathepsin B1 from Schistosoma mansoni | | Descriptor: | ACETATE ION, CHLORIDE ION, Cathepsin B-like peptidase (C01 family) | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2012-11-16 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Activation route of the Schistosoma mansoni cathepsin B1 drug target: structural map with a glycosaminoglycan switch
Structure, 22, 2014
|
|
4Z1N
 
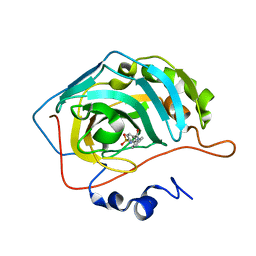 | | Carbonic anhydrase inhibitors: Design and synthesis of new heteroaryl-N-carbonylbenzenesulfonamides targeting druggable human carbonic anhydrase isoforms (hCA VII, hCA IX, and hCA XIV) | | Descriptor: | 4-[(6,7-dimethoxy-3,4-dihydro-1H-isoquinolin-2-yl)carbonyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Pospisilova, K, Rezacova, P, Pachl, P. | | Deposit date: | 2015-03-27 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Carbonic anhydrase inhibitors: Design, synthesis and structural characterization of new heteroaryl-N-carbonylbenzenesulfonamides targeting druggable human carbonic anhydrase isoforms.
Eur.J.Med.Chem., 102, 2015
|
|
4Z7X
 
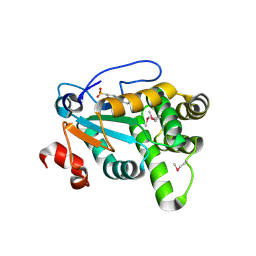 | | MdbA protein, a thiol-disulfide oxidoreductase from Actinomyces oris. | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, MdbA | | Authors: | OSIPIUK, J, Reardon-Robinson, M.E, Ton-That, H, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-04-08 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Disulfide Bond-forming Machine Is Linked to the Sortase-mediated Pilus Assembly Pathway in the Gram-positive Bacterium Actinomyces oris.
J.Biol.Chem., 290, 2015
|
|
1HG9
 
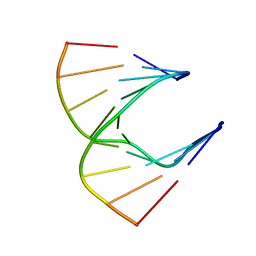 | | Solution structure of DNA:RNA hybrid | | Descriptor: | 5- D(*CP*TP*GP*AP*TP*AP*TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-13 | | Release date: | 2002-01-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of LNA:RNA duplexes by NMR: conformations and implications for RNase H activity.
Chemistry, 6, 2000
|
|
4Z5H
 
 | | HipB(S29A)-O2 20mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*AP*TP*CP*CP*TP*CP*AP*CP*TP*AP*AP*AP*GP*GP*AP*TP*AP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
4Z7Q
 
 | | Integrin alphaIIbbeta3 in complex with AGDV-NH2 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lin, F.-Y, Zhu, J, Springer, T.A. | | Deposit date: | 2015-04-07 | | Release date: | 2015-12-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | beta-Subunit Binding Is Sufficient for Ligands to Open the Integrin alpha IIb beta 3 Headpiece.
J.Biol.Chem., 291, 2016
|
|
8F93
 
 | | WDR5 covalently modified at Y228 by (R)-2-SF | | Descriptor: | 3-ethynyl-5-{[(3R)-4-{1-[(2-methoxyphenyl)methyl]-1H-benzimidazole-5-carbonyl}-3-methylpiperazin-1-yl]methyl}benzene-1-sulfonyl fluoride, CHLORIDE ION, GLYCEROL, ... | | Authors: | Taunton, J, Craven, G.B, Chen, Y. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Direct mapping of ligandable tyrosines and lysines in cells with chiral sulfonyl fluoride probes.
Nat.Chem., 15, 2023
|
|
2BEB
 
 | | X-ray structure of Asn to Thr mutant of Winged Bean Chymotrypsin inhibitor | | Descriptor: | Chymotrypsin inhibitor 3 | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-24 | | Release date: | 2006-06-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
