4L5B
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the DI-43 inhibitor | | Descriptor: | 1-[5-(4-{[(4,6-diaminopyrimidin-2-yl)sulfanyl]methyl}-5-propyl-1,3-thiazol-2-yl)-2-methoxyphenoxy]-2-methylpropan-2-ol, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2013-06-10 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Development of new deoxycytidine kinase inhibitors and noninvasive in vivo evaluation using positron emission tomography.
J.Med.Chem., 56, 2013
|
|
6FQO
 
 | | Crystal structure of CREBBP bromodomain complexd with DT29 | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, ~{N}-[3-(2,5-dimethyl-3-oxidanylidene-1,2-oxazol-4-yl)-5-(5-ethanoyl-2-ethoxy-phenyl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
6FQU
 
 | | Crystal structure of CREBBP bromodomain complexd with DR09 | | Descriptor: | 1-[3-[3-[3,3-bis(fluoranyl)piperidin-1-yl]phenyl]-4-ethoxy-phenyl]ethanone, CREB-binding protein | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
6FTF
 
 | | Regulatory subunit of a cAMP-independent protein kinase A from Trypanosoma cruzi at 1.09 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 7-cyano-7-deazainosine, Protein kinase A regulatory subunit, ... | | Authors: | Volpato Santos, Y, Lorentzen, E, Basquin, J, Boshart, M. | | Deposit date: | 2018-02-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.09151936 Å) | | Cite: | Nucleoside analogue activators of cyclic AMP-independent protein kinase A of Trypanosoma.
Nat Commun, 10, 2019
|
|
1CV2
 
 | | Hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis UT26 AT 1.6 A resolution | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Marek, J, Vevodova, J, Damborsky, J, Smatanova, I, Svensson, L.A, Newman, J, Nagata, Y, Takagi, M. | | Deposit date: | 1999-08-22 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26.
Biochemistry, 39, 2000
|
|
4E3A
 
 | | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42 | | Descriptor: | ADENOSINE, sugar kinase protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-09 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | CRYSTAL STRUCTURE OF probable sugar kinase protein from Rhizobium etli CFN 42
To be Published
|
|
6GAX
 
 | |
1RC5
 
 | |
1A6N
 
 | | DEOXY-MYOGLOBIN, ATOMIC RESOLUTION | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Vojtechovsky, J, Chu, K, Berendzen, J, Sweet, R.M, Schlichting, I. | | Deposit date: | 1998-02-26 | | Release date: | 1999-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of myoglobin-ligand complexes at near-atomic resolution.
Biophys.J., 77, 1999
|
|
6FZH
 
 | | Crystal structure of a streptococcal dehydrogenase at 1.5 Angstroem resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Gomez, S, Querol-Garcia, J, Sanchez-Barron, G, Subias, M, Gonzalez-Alsina, A, Melchor-Tafur, C, Franco-Hidalgo, V, Alberti, S, Rodriguez de Cordoba, S, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2018-03-14 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Antimicrobials Anacardic Acid and Curcumin Are Not-Competitive Inhibitors of Gram-Positive Bacterial Pathogenic Glyceraldehyde-3-Phosphate Dehydrogenase by a Mechanism Unrelated to Human C5a Anaphylatoxin Binding.
Front Microbiol, 10, 2019
|
|
3B6V
 
 | | Crystal structure of the motor domain of human kinesin family member 3C in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF3C, MAGNESIUM ION, ... | | Authors: | Zhu, H, Tempel, W, Shen, Y, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Motor domain of human kinesin family member 3C in complex with ADP.
To be Published
|
|
3B84
 
 | | Crystal structure of the human BTB domain of the Krueppel related Zinc Finger Protein 3 (HKR3) | | Descriptor: | 1,2-ETHANEDIOL, UNKNOWN ATOM OR ION, Zinc finger and BTB domain-containing protein 48 | | Authors: | Filippakopoulos, P, Bullock, A, Cooper, C, Keates, T, Salah, E, Pilka, E, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-31 | | Release date: | 2007-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of the Human BTB domain of the Krueppel related Zinc Finger Protein 3 (HKR3).
To be Published
|
|
1R9Q
 
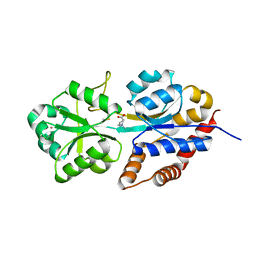 | | structure analysis of ProX in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, Glycine betaine-binding periplasmic protein, UNKNOWN ATOM OR ION | | Authors: | Schiefner, A, Breed, J, Bosser, L, Kneip, S, Gade, J, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cation-pi Interactions as Determinants for Binding of the Compatible Solutes Glycine Betaine and Proline Betaine by the Periplasmic Ligand-binding Protein ProX from Escherichia coli
J.BIOL.CHEM., 279, 2004
|
|
7L15
 
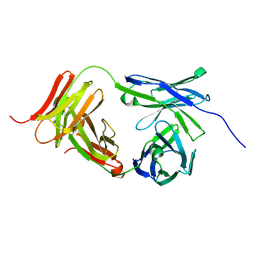 | | Marsupial T cell receptor Spl_118 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, T cell receptor gamma chain, ... | | Authors: | Wegrecki, M, Kannan Sivaraman, K, Praveena, T, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The molecular assembly of the marsupial gamma mu T cell receptor defines a third T cell lineage.
Science, 371, 2021
|
|
4GP3
 
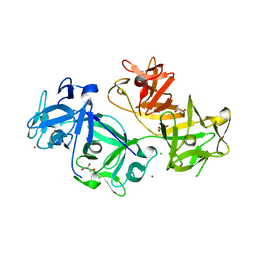 | | The crystal structure of human fascin 1 K358A mutant | | Descriptor: | BROMIDE ION, CHLORIDE ION, Fascin, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
3SRX
 
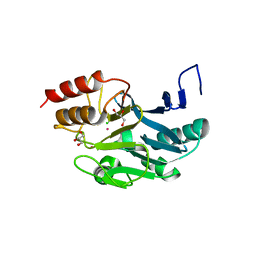 | | New Delhi Metallo-beta-Lactamase-1 Complexed with Cd | | Descriptor: | Beta-lactamase NDM-1, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-07-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New Delhi Metallo-beta-Lactamase-1 Complexed with Cd
To be Published
|
|
7KP1
 
 | | CD1a-42:2 SM binary complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, T-cell surface glycoprotein CD1a, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | CD1a selectively captures endogenous cellular lipids that broadly block T cell response.
J.Exp.Med., 218, 2021
|
|
5G4Z
 
 | | Structural basis for carboxylic acid recognition by a Cache chemosensory domain. | | Descriptor: | Methyl-accepting chemotaxis sensory transducer with Cache sensor, TRIETHYLENE GLYCOL, UNKNOWN LIGAND | | Authors: | Brewster, J, McKellar, J.L.O, Newman, J, Peat, T.S, Gerth, M.L. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for ligand recognition by a Cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae.
Sci Rep, 6, 2016
|
|
3B3N
 
 | | Structure of neuronal NOS heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-aminopyridin-2"-yl)methyl]pyrrolidin-3'-yl}ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4S)-4-[(6-aminopyridin-2-yl)methyl]pyrrolidin-3-yl}ethane-1,2-diamine, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Minimal pharmacophoric elements and fragment hopping, an approach directed at molecular diversity and isozyme selectivity. Design of selective neuronal nitric oxide synthase inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
7KP0
 
 | | CD1a-42:1 SM binary complex | | Descriptor: | (4R,7S)-4-hydroxy-7-[(1S,2E)-1-hydroxyhexadec-2-en-1-yl]-N,N,N-trimethyl-4,9-dioxo-3,5-dioxa-8-aza-4lambda~5~-phosphadotriacontan-1-aminium, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD1a selectively captures endogenous cellular lipids that broadly block T cell response.
J.Exp.Med., 218, 2021
|
|
1BVI
 
 | | RIBONUCLEASE T1 (WILDTYPE) COMPLEXED WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Langhorst, U, Loris, R, Denisov, V.P, Doumen, J, Roose, P, Maes, D, Halle, B, Steyaert, J. | | Deposit date: | 1998-09-15 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissection of the structural and functional role of a conserved hydration site in RNase T1.
Protein Sci., 8, 1999
|
|
7KOZ
 
 | | CD1a-36:2 SM binary complex | | Descriptor: | (4S,7S,17Z)-4-hydroxy-7-[(1S,2E)-1-hydroxyhexadec-2-en-1-yl]-N,N,N-trimethyl-4,9-dioxo-3,5-dioxa-8-aza-4lambda~5~-phosphahexacos-17-en-1-aminium, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CD1a selectively captures endogenous cellular lipids that broadly block T cell response.
J.Exp.Med., 218, 2021
|
|
4H5S
 
 | | Complex structure of Necl-2 and CRTAM | | Descriptor: | Cell adhesion molecule 1, Cytotoxic and regulatory T-cell molecule | | Authors: | Zhang, S, Lu, G, Qi, J, Li, Y, Zhang, Z, Zhang, B, Yan, J, Gao, G.F. | | Deposit date: | 2012-09-18 | | Release date: | 2013-08-07 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Competition of cell adhesion and immune recognition: insights into the interaction between CRTAM and nectin-like 2.
Structure, 21, 2013
|
|
4GP0
 
 | | The crystal structure of human fascin 1 R149A K150A R151A mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
5TT7
 
 | | Discovery of TAK-659, an Orally Available Investigational Inhibitor of Spleen Tyrosine Kinase (SYK) | | Descriptor: | 2-{[(1R,2S)-2-aminocyclohexyl]amino}-4-[(3-methylphenyl)amino]-6,7-dihydro-5H-pyrrolo[3,4-d]pyrimidin-5-one, Tyrosine-protein kinase SYK | | Authors: | Yano, J, Jennings, A, Lam, B, Hoffman, I.D. | | Deposit date: | 2016-11-01 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery of TAK-659 an orally available investigational inhibitor of Spleen Tyrosine Kinase (SYK).
Bioorg. Med. Chem. Lett., 26, 2016
|
|
