3JAD
 
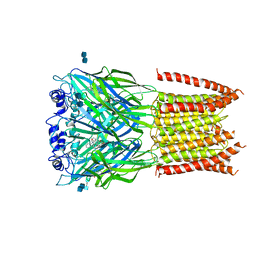 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, strychnine-bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1, STRYCHNINE | | Authors: | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
3JQJ
 
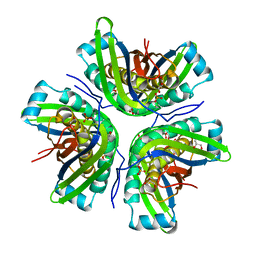 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 | | Descriptor: | GLYCEROL, Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION, ... | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3JR2
 
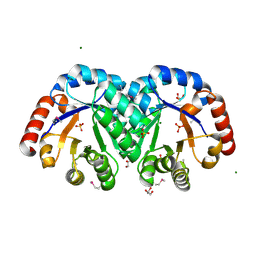 | | X-ray crystal structure of the Mg-bound 3-keto-L-gulonate-6-phosphate decarboxylase from Vibrio cholerae O1 biovar El Tor str. N16961 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hexulose-6-phosphate synthase SgbH, ... | | Authors: | Nocek, B, Maltseva, N, Stam, J, Anderson, W, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Mg-bound 3-keto-L-gulonate-6-phosphate
decarboxylase from Vibrio cholerae O1 biovar El Tor str. N16961
To be Published
|
|
3JAI
 
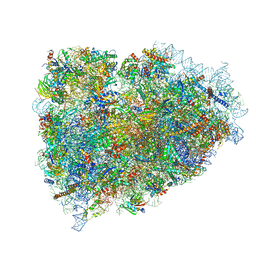 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UGA stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
3ULU
 
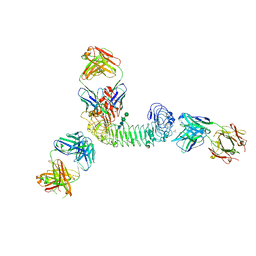 | | Structure of quaternary complex of human TLR3ecd with three Fabs (Form1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1068 heavy chain, ... | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
3JS2
 
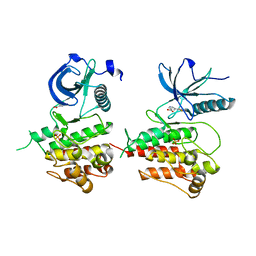 | | Crystal structure of minimal kinase domain of fibroblast growth factor receptor 1 in complex with 5-(2-thienyl)nicotinic acid | | Descriptor: | 5-(2-thienyl)nicotinic acid, Basic fibroblast growth factor receptor 1, PHOSPHATE ION | | Authors: | Bae, J.H, Ravindranathan, K.P, Mandiyan, V, Ekkati, A.R, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2009-09-09 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of novel fibroblast growth factor receptor 1 kinase inhibitors by structure-based virtual screening
J.Med.Chem., 53, 2010
|
|
3USH
 
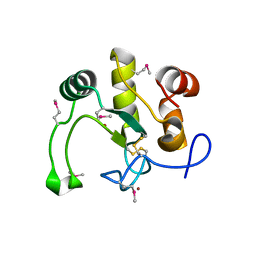 | | Crystal Structure of the Q2S0R5 protein from Salinibacter ruber, Northeast Structural Genomics Consortium Target SrR207 | | Descriptor: | BROMIDE ION, Uncharacterized protein | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Crystal Structure of the Q2S0R5 protein from Salinibacter ruber, Northeast Structural Genomics Consortium Target SrR207
To be Published
|
|
3USO
 
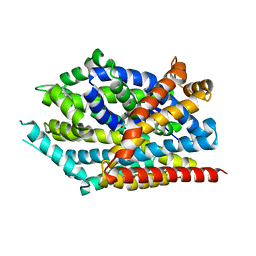 | |
3JVN
 
 | | Crystal Structure of the acetyltransferase VF_1542 from Vibrio fischeri, Northeast Structural Genomics Consortium Target VfR136 | | Descriptor: | Acetyltransferase, SULFATE ION | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-17 | | Release date: | 2009-09-29 | | Last modified: | 2019-08-07 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Northeast Structural Genomics Consortium Target VfR136
To be Published
|
|
3UTT
 
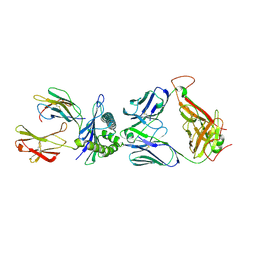 | | 1E6-A*0201-ALWGPDPAAA Complex, Triclinic | | Descriptor: | 1E6 TCR Alpha Chain, 1E6 TCR Beta Chain, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Bulek, A.M, Rossjohn, J, Gras, S. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-25 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the killing of human beta cells by CD8(+) T cells in type 1 diabetes.
Nat.Immunol., 13, 2012
|
|
3UVF
 
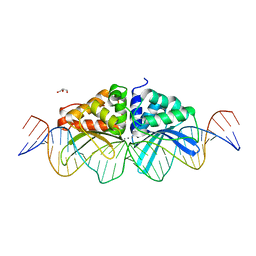 | | Expanding LAGALIDADG endonuclease scaffold diversity by rapidly surveying evolutionary sequence space | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Intron-encoded DNA endonuclease I-HjeMI, ... | | Authors: | Jacoby, K, Metzger, M, Shen, B, Jarjour, J, Stoddard, B, Scharenberg, A. | | Deposit date: | 2011-11-29 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Expanding LAGLIDADG endonuclease scaffold diversity by rapidly surveying evolutionary sequence space.
Nucleic Acids Res., 40, 2012
|
|
3UZ0
 
 | |
3UJJ
 
 | |
3K4A
 
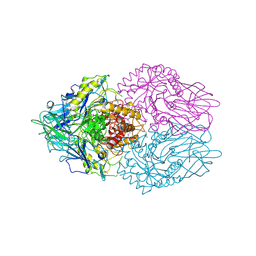 | |
1BH1
 
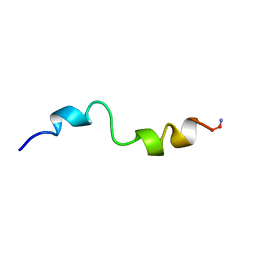 | | STRUCTURAL STUDIES OF D-PRO MELITTIN, NMR, 20 STRUCTURES | | Descriptor: | MELITTIN | | Authors: | Barnham, K.J, Hewish, D, Werkmeister, J, Curtain, C, Kirkpatrick, A, Bartone, N, Liu, S.T, Norton, R, Rivett, D. | | Deposit date: | 1998-06-11 | | Release date: | 1999-01-06 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure and activity of D-Pro14 melittin.
J.Protein Chem., 21, 2002
|
|
3K6Y
 
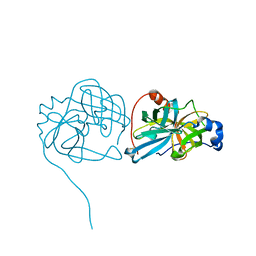 | | Crystal structure of Rv3671c protease from M. tuberculosis, active form | | Descriptor: | POSSIBLE MEMBRANE-ASSOCIATED SERINE PROTEASE | | Authors: | Biswas, T, Small, J, Vandal, O, Ehrt, S, Tsodikov, O.V. | | Deposit date: | 2009-10-10 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insight into serine protease Rv3671c that Protects M. tuberculosis from oxidative and acidic stress.
Structure, 18, 2010
|
|
3K7V
 
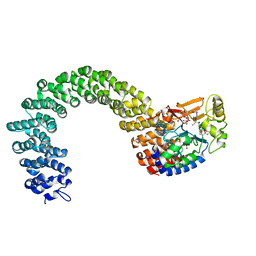 | | Protein phosphatase 2A core complex bound to dinophysistoxin-1 | | Descriptor: | (2R)-3-[(2S,5R,6R,8S)-8-{(1R,2E)-3-[(2R,4a'R,5R,6'S,8'R,8a'S)-6'-{(1S,3S)-3-[(2S,3R,6R,11R)-3,11-dimethyl-1,7-dioxaspiro[5.5]undec-2-yl]-1-hydroxybutyl}-8'-hydroxy-7'-methylideneoctahydro-3H,3'H-spiro[furan-2,2'-pyrano[3,2-b]pyran]-5-yl]-1-methylprop-2-en-1-yl}-5-hydroxy-10-methyl-1,7-dioxaspiro[5.5]undec-10-en-2-yl]-2-hydroxy-2-methylpropanoic acid, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Jeffrey, P.D, Huhn, J, Shi, Y. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A structural basis for the reduced toxicity of dinophysistoxin-2.
Chem.Res.Toxicol., 22, 2009
|
|
3K2E
 
 | |
3K96
 
 | | 2.1 Angstrom resolution crystal structure of glycerol-3-phosphate dehydrogenase (gpsA) from Coxiella burnetii | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, Glycerol-3-phosphate dehydrogenase [NAD(P)+] | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of Glycerol-3-phosphate Dehydrogenase (gpsA) from Coxiella burnetii.
TO BE PUBLISHED
|
|
3UPL
 
 | | Crystal structure of the Brucella abortus enzyme catalyzing the first committed step of the methylerythritol 4-phosphate pathway. | | Descriptor: | GLYCEROL, MAGNESIUM ION, Oxidoreductase | | Authors: | Calisto, B.M, Perez-Gil, J, Fita, I, Rodriguez-Concepcion, M. | | Deposit date: | 2011-11-18 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Brucella abortus deoxyxylulose-5-phosphate reductoisomerase-like (DRL) enzyme involved in isoprenoid biosynthesis.
J.Biol.Chem., 287, 2012
|
|
3V72
 
 | | Crystal Structure of Rat DNA polymerase beta Mutator E295K: Enzyme-dsDNA | | Descriptor: | CHLORIDE ION, DNA 5'-D(P*AP*AP*AP*CP*TP*CP*AP*CP*AP*T)-3', DNA 5'-D(P*AP*TP*GP*TP*GP*AP*GP*T)-3', ... | | Authors: | Gridley, C.L, Jaeger, J. | | Deposit date: | 2011-12-20 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Unfavorable Electrostatic and Steric Interactions in DNA Polymerase beta E295K Mutant Interfere with the Enzyme s Pathway
J.Am.Chem.Soc., 134, 2012
|
|
3USI
 
 | |
3V7B
 
 | | Dip2269 protein from corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein | | Authors: | Osipiuk, J, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-11 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | Dip2269 protein from corynebacterium diphtheriae.
To be Published
|
|
3ULV
 
 | | Structure of quaternary complex of human TLR3ecd with three Fabs (Form2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1068 heavy chain, ... | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.522 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
3J3I
 
 | | Penicillium chrysogenum virus (PcV) capsid structure | | Descriptor: | Capsid protein | | Authors: | Luque, D, Gomez-Blanco, J, Garriga, D, Brilot, A, Gonzalez, J.M, Havens, W.H, Carrascosa, J.L, Trus, B.L, Verdaguer, N, Grigorieff, N, Ghabrial, S.A, Caston, J.R. | | Deposit date: | 2013-03-08 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM near-atomic structure of a dsRNA fungal virus shows ancient structural motifs preserved in the dsRNA viral lineage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
