2DUV
 
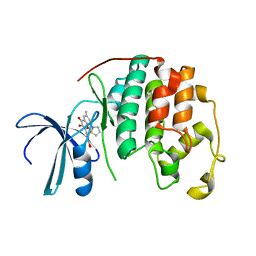 | | Structure of CDK2 with a 3-hydroxychromones | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)-8-(1,1-DIOXIDOISOTHIAZOLIDIN-2-YL)-3-HYDROXY-6-METHYL-4H-CHROMEN-4-ONE, Cell division protein kinase 2 | | Authors: | Kim, K.H, Lee, J, Park, T, Jeong, S, Hong, C. | | Deposit date: | 2006-07-27 | | Release date: | 2007-01-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3-Hydroxychromones as cyclin-dependent kinase inhibitors: synthesis and biological evaluation.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5V34
 
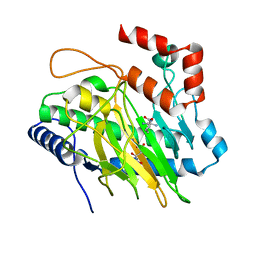 | | Ethylene forming enzyme in complex with manganese, malic acid and L-arginine | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININE, D-MALATE, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
3A81
 
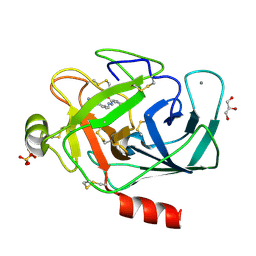 | |
2DFS
 
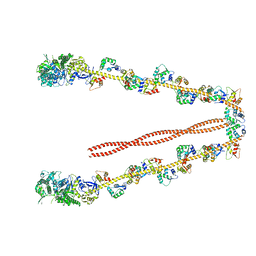 | | 3-D structure of Myosin-V inhibited state | | Descriptor: | Calmodulin, Myosin-5A | | Authors: | Liu, J, Taylor, D.W, Krementsova, E.B, Trybus, K.M, Taylor, K.A. | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (24 Å) | | Cite: | Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography
Nature, 442, 2006
|
|
3AH5
 
 | | Crystal Structure of flavin dependent thymidylate synthase ThyX from helicobacter pylori complexed with FAD and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Zhang, X, Zhang, J, Hu, Y, Zou, Q, Wang, D. | | Deposit date: | 2010-04-14 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and functional analysis of a flavin dependent thymidylate synthase from helicobacter pylori
To be Published
|
|
5V2U
 
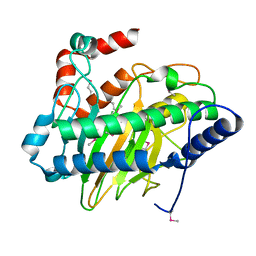 | | Ethylene forming enzyme apo form | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.058 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
5V32
 
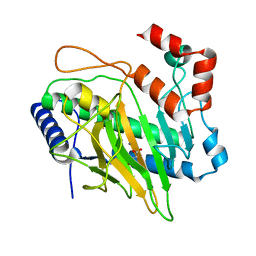 | | Ethylene forming enzyme in complex with manganese and malic acid | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, D-MALATE, MANGANESE (II) ION | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
2DPE
 
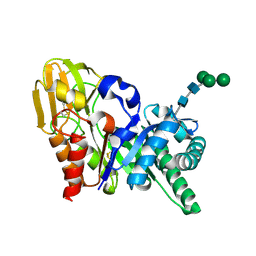 | | Crystal structure of a secretory 40KDA glycoprotein from sheep at 2.0A resolution | | Descriptor: | Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Singh, N, Sharma, S, Das, U, Srinivasan, A, Singh, T.P. | | Deposit date: | 2006-05-11 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of a secretory signalling glycoprotein from sheep at 2.0A resolution
J.Struct.Biol., 156, 2006
|
|
3ALW
 
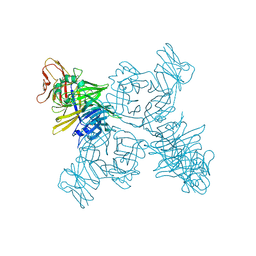 | | Crystal structure of the measles virus hemagglutinin bound to its cellular receptor SLAM (Form I, MV-H-SLAM(N102H/R108Y) fusion) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, CDw150 | | Authors: | Hashiguchi, T, Ose, T, Kubota, M, Maita, N, Kamishikiryo, J, Maenaka, K, Yanagi, Y. | | Deposit date: | 2010-08-09 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM
Nat.Struct.Mol.Biol., 18, 2011
|
|
1QF9
 
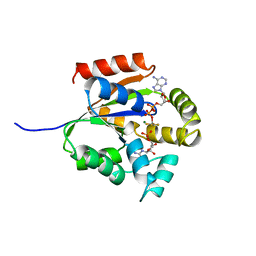 | |
2DSD
 
 | |
1CZ4
 
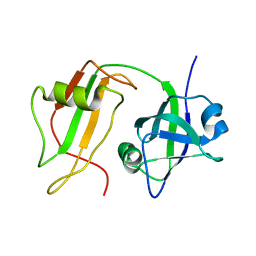 | | NMR STRUCTURE OF VAT-N: THE N-TERMINAL DOMAIN OF VAT (VCP-LIKE ATPASE OF THERMOPLASMA) | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Coles, M, Diercks, T, Liermann, J, Groeger, A, Rockel, B, Baumeister, W, Koretke, K, Lupas, A, Peters, J, Kessler, H. | | Deposit date: | 1999-09-01 | | Release date: | 1999-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of VAT-N reveals a 'missing link' in the evolution of complex enzymes from a simple betaalphabetabeta element.
Curr.Biol., 9, 1999
|
|
1NDK
 
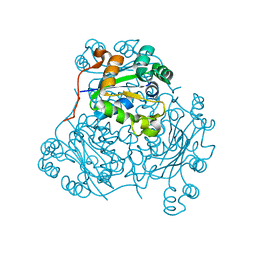 | | X-RAY STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Dumas, C, Morera, S, Lascu, I, Veron, M. | | Deposit date: | 1993-07-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of nucleoside diphosphate kinase.
EMBO J., 11, 1992
|
|
5UNO
 
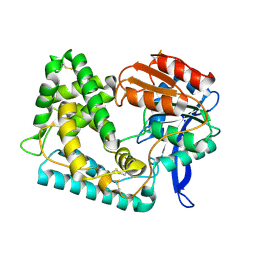 | | Crystal Structure of Hip1 (Rv2224c) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-12 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
2DR2
 
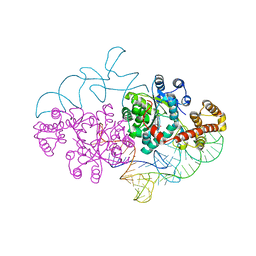 | | Structure of human tryptophanyl-tRNA synthetase in complex with tRNA(Trp) | | Descriptor: | SULFATE ION, TRYPTOPHAN, Tryptophanyl-tRNA synthetase, ... | | Authors: | Shen, N, Guo, L, Yang, B, Jin, Y, Ding, J. | | Deposit date: | 2006-06-05 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human tryptophanyl-tRNA synthetase in complex with tRNA(Trp) reveals the molecular basis of tRNA recognition and specificity
Nucleic Acids Res., 34, 2006
|
|
5V31
 
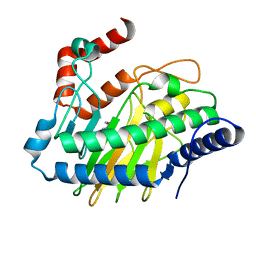 | | Ethylene forming enzyme in complex with manganese and L-arginine | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININE, MANGANESE (II) ION | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
2DT1
 
 | | Crystal Structure Of The Complex Of Goat Signalling Protein With Tetrasaccharide At 2.09 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, J, Ethayathulla, A.S, Srivastava, D.B, Singh, N, Sharma, S, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2006-07-09 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Carbohydrate-binding properties of goat secretory glycoprotein (SPG-40) and its functional implications: structures of the native glycoprotein and its four complexes with chitin-like oligosaccharides
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
3A61
 
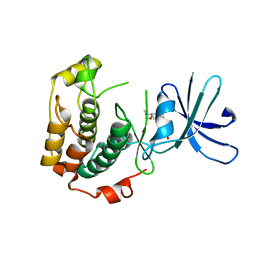 | | Crystal structure of unphosphorylated p70S6K1 (Form II) | | Descriptor: | Ribosomal protein S6 kinase beta-1, STAUROSPORINE | | Authors: | Sunami, T, Byrne, N, Diehl, R.E, Funabashi, K, Hall, D.L, Ikuta, M, Patel, S.B, Shipman, J.M, Smith, R.F, Takahashi, I, Zugay-Murphy, J, Iwasawa, Y, Lumb, K.J, Munshi, S.K, Sharma, S. | | Deposit date: | 2009-08-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Structural basis of human p70 ribosomal S6 kinase-1 regulation by activation loop phosphorylation.
J.Biol.Chem., 285, 2010
|
|
4GQM
 
 | |
3A7Y
 
 | |
5V9T
 
 | | Crystal structure of selective pyrrolidine amide KDM5a inhibitor N-{(3R)-1-[3-(propan-2-yl)-1H-pyrazole-5-carbonyl]pyrrolidin-3-yl}cyclopropanecarboxamide (compound 48) | | Descriptor: | Lysine-specific demethylase 5A, N-{(3R)-1-[3-(propan-2-yl)-1H-pyrazole-5-carbonyl]pyrrolidin-3-yl}cyclopropanecarboxamide, NICKEL (II) ION, ... | | Authors: | Kiefer, J.R, Liang, J, Vinogradova, M. | | Deposit date: | 2017-03-23 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | From a novel HTS hit to potent, selective, and orally bioavailable KDM5 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3A8A
 
 | |
2DPK
 
 | |
1NAY
 
 | | GPP-Foldon:X-ray structure | | Descriptor: | Collagen-like peptide | | Authors: | Stetefeld, J. | | Deposit date: | 2002-11-29 | | Release date: | 2003-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Collagen Stabilization at Atomic Level. Crystal Structure of Designed (GlyProPro)(10)foldon
Structure, 11, 2003
|
|
3ALL
 
 | | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, mutant Y270A | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kobayashi, J, Yoshida, H, Yoshikane, Y, Kamitori, S, Yagi, T. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be published
|
|
