8C2H
 
 | | Transmembrane domain of active state homomeric GluA1 AMPA receptor in tandem with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
5EWD
 
 | |
8C1P
 
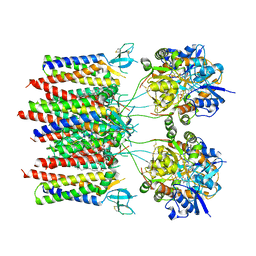 | | Active state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CYCLOTHIAZIDE, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
4HOW
 
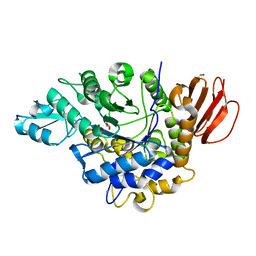 | | The crystal structure of isomaltulose synthase from Erwinia rhapontici NX5 | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-22 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
8C9D
 
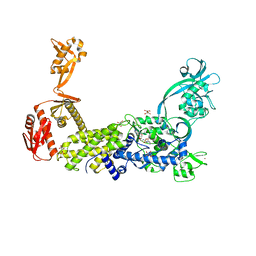 | | Priestia megaterium W130Q mutant of type 2 isoleucyl-tRNA synthetase complexed with an isoleucyl-adenylate analogue | | Descriptor: | D(-)-TARTARIC ACID, Isoleucine--tRNA ligase, N-[ISOLEUCINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
4HPH
 
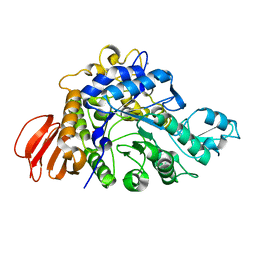 | | The crystal structure of isomaltulose synthase mutant E295Q from Erwinia rhapontici NX5 in complex with its natural substrate sucrose | | Descriptor: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | Authors: | Xu, Z, Li, S, Xu, H, Zhou, J. | | Deposit date: | 2012-10-23 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
8C5U
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 8-mer RNA, pppGpGpUpApApApUpG (IC8) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-01-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
8C9F
 
 | | Priestia megaterium inactive HIGH motif mutant of type1 isoleucyl-tRNA synthetase complexed with an isoleucyl-adenylate analogue. | | Descriptor: | CHLORIDE ION, Isoleucine--tRNA ligase, LITHIUM ION, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
8C1S
 
 | | Transmembrane domain of resting state homomeric GluA2 F231A mutant AMPA receptor in complex with TARP gamma 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C2I
 
 | | Transmembrane domain of resting state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C1R
 
 | | Resting state homomeric GluA2 F231A mutant AMPA receptor in complex with TARP gamma-2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
5F02
 
 | | CATHEPSIN L IN COMPLEX WITH (2S,4R)-4-(2-Chloro-4-methoxy-benzenesulfonyl)-1-[3-(5-chloro-pyridin-2-yl)-azetidine-3-carbonyl]-pyrrolidine-2-carboxylic acid (1-cyano-cyclopropyl)-amide | | Descriptor: | (2~{S},4~{R})-4-[(2-chloranyl-4-methoxy-phenyl)-bis(oxidanyl)-$l^{4}-sulfanyl]-1-[3-(5-chloranylpyridin-2-yl)azetidin-3-yl]carbonyl-~{N}-[1-(iminomethyl)cyclopropyl]pyrrolidine-2-carboxamide, Cathepsin L1, GLYCEROL | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
8C1Q
 
 | | Resting state homomeric GluA1 AMPA receptor in complex with TARP gamma 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Ivica, J, Krieger, J.M, Ho, H, Yamashita, K, Cais, O, Greger, I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8C5S
 
 | | Cryo-EM structure of yeast mitochondrial RNA polymerase transcription initiation complex with 7-mer RNA, pppGpGpUpApApApU (IC7) | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Goovaerts, Q, Shen, J, Patel, S.S, Das, K. | | Deposit date: | 2023-01-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structures illustrate step-by-step mitochondrial transcription initiation.
Nature, 622, 2023
|
|
5F00
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH 5-[3-[(3-chloro-8-quinolyl)amino]phenyl]-5-methyl-2,6-dihydro-1,4-oxazin-3-amine | | Descriptor: | (5~{R})-5-[3-[(3-chloranylquinolin-8-yl)amino]phenyl]-5-methyl-2,6-dihydro-1,4-oxazin-3-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
8C8V
 
 | | Priestia megaterium mupirocin-resistant isoleucyl-tRNA synthetase 2 with a fully-resolved C-terminal tRNA-binding domain complexed with an isoleucyl-adenylate analogue | | Descriptor: | D(-)-TARTARIC ACID, Isoleucine--tRNA ligase, N-[ISOLEUCINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
8C9E
 
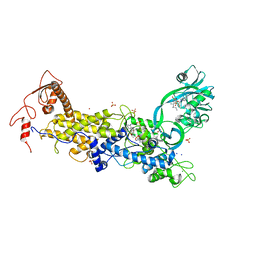 | | Priestia megaterium mupirocin-sensitive isoleucyl-tRNA synthetase 1 complexed with an isoleucyl-adenylate analogue | | Descriptor: | CHLORIDE ION, Isoleucine--tRNA ligase, LITHIUM ION, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
8C54
 
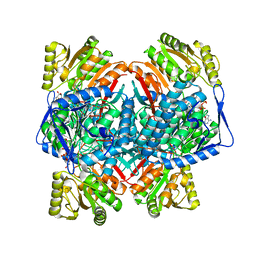 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | Authors: | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
1AQC
 
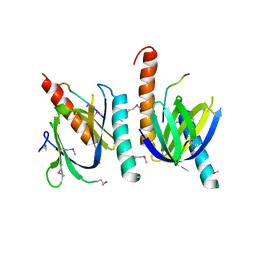 | | X11 PTB DOMAIN-10MER PEPTIDE COMPLEX | | Descriptor: | PEPTIDE, X11 | | Authors: | Lee, C.-H, Zhang, Z, Kuriyan, J. | | Deposit date: | 1997-07-28 | | Release date: | 1997-12-24 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain.
EMBO J., 16, 1997
|
|
5F01
 
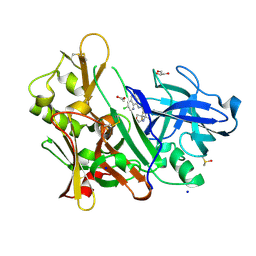 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH (1SR,2SR)-2-((R)-2-amino-5,5-difluoro-4-methyl-5,6-dihydro-4H-1,3-oxazin-4-yl)-N-(3-chloroquinolin-8-yl)cyclopropanecarboxamide | | Descriptor: | (1~{R},2~{R})-2-[(4~{R})-2-azanyl-5,5-bis(fluoranyl)-4-methyl-6~{H}-1,3-oxazin-4-yl]-~{N}-(3-chloranylquinolin-8-yl)cyclopropane-1-carboxamide, ACETATE ION, Beta-secretase 1, ... | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
8CAP
 
 | |
5EZX
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH {(1R,2R)-2-[(R)-2-Amino-4-(4-difluoromethoxy-phenyl)-4,5-dihydro-oxazol-4-yl]-cyclopropyl}-(5-chloro-pyridin-3-yl)-methanone | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Banner, D, Benz, J, Stihle, M, Kuglstatter, A. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-24 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
8CAL
 
 | |
8CMN
 
 | |
8CAO
 
 | | Crystal structure of dehydrogenase domain of Cylindrospermum stagnale NADPH-Oxidase 5 (NOX5) in complex with CA24 | | Descriptor: | 1,2-ETHANEDIOL, 8-[2-(4-cyclohexylphenyl)quinolin-4-yl]carbonyl-1,3,8-triazaspiro[4.5]decane-2,4-dione, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reis, J, Mattevi, A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting ROS production through inhibition of NADPH oxidases.
Nat.Chem.Biol., 19, 2023
|
|
