7OVD
 
 | | Human soluble adenylyl cyclase in complex with the inhibitor TDI10229 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-6-[1,5-dimethyl-4-(phenylmethyl)pyrazol-3-yl]pyrimidin-2-amine, ACETATE ION, ... | | Authors: | Steegborn, C, Quast, J. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of TDI-10229: A Potent and Orally Bioavailable Inhibitor of Soluble Adenylyl Cyclase (sAC, ADCY10).
Acs Med.Chem.Lett., 12, 2021
|
|
8PXS
 
 | | Short RNA binding to peptide amyloids | | Descriptor: | RNA (5'-R(P*GP*UP*CP*A)-3'), VAL-ALA-GLN-ALA-GLN-ILE-ASN-ILE | | Authors: | Rout, S.K, Cadalbert, R, Schroder, N, Wiegand, T, Zehnder, J, Gampp, O, Guntert, P, Kringler, D, Kreutz, C, Knorlein, A, Hall, J, Greenwald, J, Riek, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-16 | | Method: | SOLID-STATE NMR | | Cite: | An Analysis of Nucleotide-Amyloid Interactions Reveals Selective Binding to Codon-Sized RNA.
J.Am.Chem.Soc., 145, 2023
|
|
6ODA
 
 | | Crystal structure of HDAC8 in complex with compound 2 | | Descriptor: | Histone deacetylase 8, N-{2-[3-(hydroxyamino)-3-oxopropyl]phenyl}-3-(trifluoromethyl)benzamide, POTASSIUM ION, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
1EOV
 
 | | FREE ASPARTYL-TRNA SYNTHETASE (ASPRS) (E.C. 6.1.1.12) FROM YEAST | | Descriptor: | ASPARTYL-TRNA SYNTHETASE | | Authors: | Sauter, C, Lorber, B, Cavarelli, J, Moras, D, Giege, R. | | Deposit date: | 2000-03-24 | | Release date: | 2000-09-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The free yeast aspartyl-tRNA synthetase differs from the tRNA(Asp)-complexed enzyme by structural changes in the catalytic site, hinge region, and anticodon-binding domain.
J.Mol.Biol., 299, 2000
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
6OCT
 
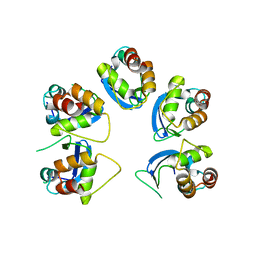 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1EH5
 
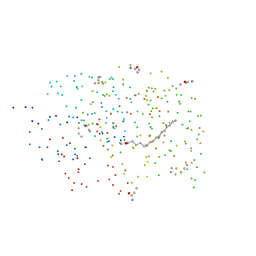 | | CRYSTAL STRUCTURE OF PALMITOYL PROTEIN THIOESTERASE 1 COMPLEXED WITH PALMITATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Bellizzi III, J.J, Widom, J, Kemp, C, Lu, J.Y, Das, A.K, Hofmann, S.L, Clardy, J. | | Deposit date: | 2000-02-18 | | Release date: | 2000-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of palmitoyl protein thioesterase 1 and the molecular basis of infantile neuronal ceroid lipofuscinosis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6ZW8
 
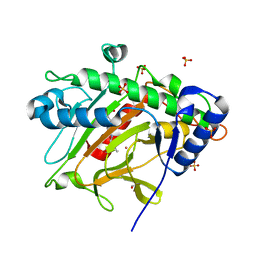 | | Isopenicillin N synthase in complex with Cd and ACV. | | Descriptor: | CADMIUM ION, GLYCEROL, Isopenicillin N synthase, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-07-27 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
7OGT
 
 | | Folded elbow of cohesin | | Descriptor: | Structural maintenance of chromosomes protein 1, Structural maintenance of chromosomes protein 3 | | Authors: | Lee, B.-G, Gonzalez Llamazares, A, Collier, J, Patele, N.J, Nasmyth, K.A, Lowe, J. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Folding of cohesin's coiled coil is important for Scc2/4-induced association with chromosomes.
Elife, 10, 2021
|
|
7OO2
 
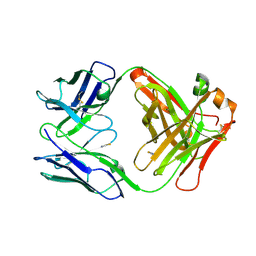 | | Crystal structure of an antibody targeting the capsular polysaccharide of serogroup X Neisseria meningitidis (MenX) | | Descriptor: | THIOCYANATE ION, anti-MenX Fab heavy chain, anti-MenX Fab light chain | | Authors: | Pietri, G.P, de Ruyck, J, Lenac, T, Adamo, R, Bouckaert, J. | | Deposit date: | 2021-05-26 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Elucidating the Structural and Minimal Protective Epitope of the Serogroup X Meningococcal Capsular Polysaccharide.
Front Mol Biosci, 8, 2021
|
|
4V94
 
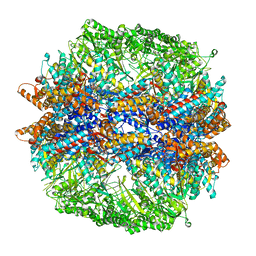 | | Molecular architecture of the eukaryotic chaperonin TRiC/CCT derived by a combination of chemical crosslinking and mass-spectrometry, XL-MS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Leitner, A, Joachimiak, L.A, Bracher, A, Walzthoeni, T, Chen, B, Monkemeyer, L, Pechmann, S, Holmes, S, Cong, Y, Ma, B, Ludtke, S, Chiu, W, Hartl, F.U, Aebersold, R, Frydman, J. | | Deposit date: | 2012-01-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Architecture of the Eukaryotic Chaperonin TRiC/CCT.
Structure, 20, 2012
|
|
4V05
 
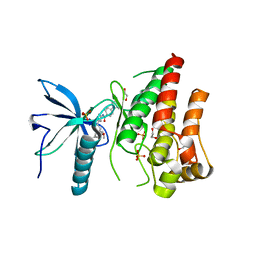 | | FGFR1 in complex with AZD4547. | | Descriptor: | 1,2-ETHANEDIOL, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, PFEIFFER SYNDROME), ... | | Authors: | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | Deposit date: | 2014-09-10 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
4UX6
 
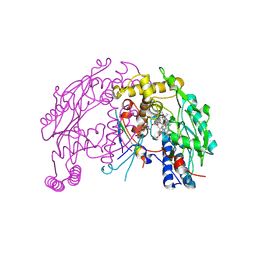 | | The discovery of novel, potent and highly selective inhibitors of inducible nitric oxide synthase (iNOS) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, INDUCIBLE, ... | | Authors: | Cheshire, D.R, Andrews, G, Beaton, H.G, Birkinshaw, T, Boughton-Smith, N, Connolly, S, Cook, T.R, Cooper, A, Cooper, S.L, Cox, D, Dixon, J, Gensmantel, N, Hamley, P.J, Harrison, R, Hartopp, P, Kack, H, Luker, T, Mete, A, Millichip, I, Nicholls, D.J, Pimm, A.D, St-Gallay, S.A, Wallace, A.V. | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Discovery of Novel, Potent and Highly Selective Inhibitors of Inducible Nitric Oxide Synthase (Inos).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1LWF
 
 | | CRYSTAL STRUCTURE OF A MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMQ+M184V: M41L/D67N/K70R/M184V/T215Y) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
6ZZ6
 
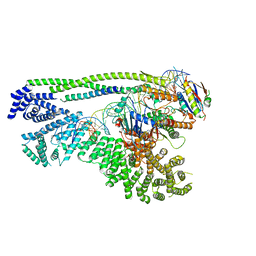 | | Cryo-EM structure of S.cerevisiae cohesin-Scc2-DNA complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (34-MER), MAGNESIUM ION, ... | | Authors: | Lee, B.-G, Gonzalez Llamazares, A, Collier, J, Nasmyth, K.A, Lowe, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Transport of DNA within cohesin involves clamping on top of engaged heads by Scc2 and entrapment within the ring by Scc3.
Elife, 9, 2020
|
|
1LW0
 
 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
4V6I
 
 | | Localization of the small subunit ribosomal proteins into a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein RACK1 (RACK1), ... | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8DGG
 
 | | Structure of glycosylated LAG-3 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Silberstein, J.L, Mathews, I.I, Frank, J.A, Chan, K.-W, Fernandez, D, Du, J, Wang, J, Kong, X.-P, Cochran, J.R. | | Deposit date: | 2022-06-23 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Structural insights reveal interplay between LAG-3 homodimerization, ligand binding, and function.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7ORB
 
 | | Crystal structure of the L452R mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-75 and COVOX-253 Fabs | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
7OR9
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and COVOX-278 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
7ORA
 
 | | Crystal structure of the T478K mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-45 and COVOX-253 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-253 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
6OHY
 
 | | Chimpanzee SIV Env trimeric ectodomain. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pallesen, J, Andrabi, R, de Val, N, Burton, D.R, Ward, A.B. | | Deposit date: | 2019-04-08 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The Chimpanzee SIV Envelope Trimer: Structure and Deployment as an HIV Vaccine Template.
Cell Rep, 27, 2019
|
|
6OG1
 
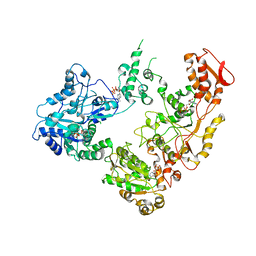 | | Focus classification structure of the hyperactive ClpB mutant K476C, bound to casein, pre-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Hyperactive disaggregase ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | Deposit date: | 2019-04-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
7OLX
 
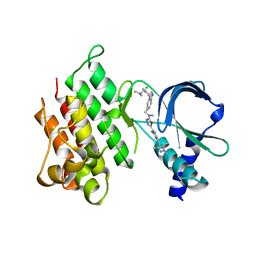 | | MerTK kinase domain with type 1.5 inhibitor containing a tri-methyl pyrazole group | | Descriptor: | CHLORIDE ION, Tyrosine-protein kinase Mer, ~{N}-[[3-[4-[(dimethylamino)methyl]phenyl]imidazo[1,2-a]pyridin-6-yl]methyl]-~{N}-methyl-5-[3-methyl-5-(1,3,5-trimethylpyrazol-4-yl)pyridin-2-yl]-1,3,4-oxadiazol-2-amine | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
7OLS
 
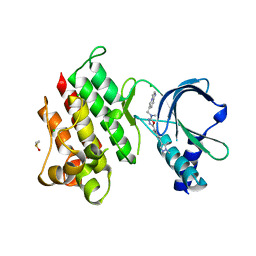 | | MerTK kinase domain with type 1.5 inhibitor containing a di-methyl pyrazole group | | Descriptor: | 5-[4-(1,5-dimethylpyrazol-4-yl)-2-methyl-phenyl]-~{N}-(imidazo[1,2-a]pyridin-6-ylmethyl)-~{N}-methyl-1,3,4-oxadiazol-2-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
