7K39
 
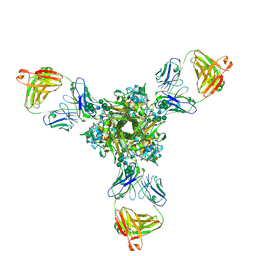 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation A, neutral pH-like | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | 著者 | Gui, M, Gao, J, Xiang, Y. | | 登録日 | 2020-09-10 | | 公開日 | 2020-11-11 | | 最終更新日 | 2020-12-09 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
1FZQ
 
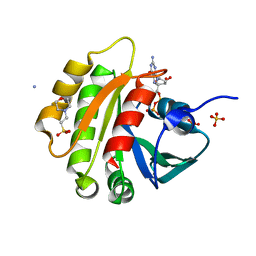 | | CRYSTAL STRUCTURE OF MURINE ARL3-GDP | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADP-RIBOSYLATION FACTOR-LIKE PROTEIN 3, AMMONIUM ION, ... | | 著者 | Hillig, R.C, Hanzal-Bayer, M, Linari, M, Becker, J, Wittinghofer, A, Renault, L. | | 登録日 | 2000-10-04 | | 公開日 | 2000-12-06 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and biochemical properties show ARL3-GDP as a distinct GTP binding protein.
Structure Fold.Des., 8, 2000
|
|
3JZY
 
 | | Crystal structure of human Intersectin 2 C2 domain | | 分子名称: | Intersectin 2, UNKNOWN ATOM OR ION | | 著者 | Shen, Y, Tempel, W, Tong, Y, Li, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | 登録日 | 2009-09-24 | | 公開日 | 2009-10-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Crystal structure of human Intersectin 2 C2 domain
To be Published
|
|
8JJK
 
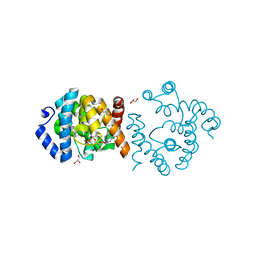 | | SP1746 in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, GLYCEROL, ... | | 著者 | Jin, Y, Niu, L, Ke, J. | | 登録日 | 2023-05-30 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 2024
|
|
8JKP
 
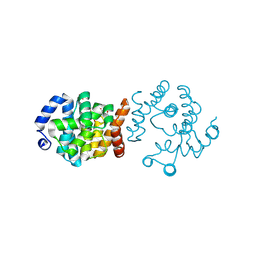 | | SP1746 in complex with dTMP | | 分子名称: | FE (III) ION, GLYCEROL, SULFATE ION, ... | | 著者 | Jin, Y, Ke, J, Niu, L. | | 登録日 | 2023-06-01 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 2024
|
|
5AEK
 
 | | Crystal structure of the human SENP2 C548S in complex with the human SUMO1 K48M F66W | | 分子名称: | SENTRIN-SPECIFIC PROTEASE 2, SMALL UBIQUITIN-RELATED MODIFIER 1 | | 著者 | Gallego, P, Grana-Montes, R, Espargaro, A, Castillo, V, Torrent, J, Lange, R, Papaleo, E, Lindorff-Larsend, K, Ventura, S, Reverter, D. | | 登録日 | 2014-12-23 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Stepping Back and Forward on Sumo Folding Evolution
To be Published
|
|
7JRN
 
 | | Crystal structure of the wild type SARS-CoV-2 papain-like protease (PLPro) with inhibitor GRL0617 | | 分子名称: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, SULFATE ION, ... | | 著者 | Sacco, M, Ma, C, Wang, J, Chen, Y. | | 登録日 | 2020-08-12 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay.
Acs Cent.Sci., 7, 2021
|
|
8J8W
 
 | | Crystal structure of AtHPPD-Y19788 complex | | 分子名称: | 2,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-phenyl-quinazolin-4-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | 著者 | Lin, H.-Y, Dong, J, Yang, G.-F. | | 登録日 | 2023-05-02 | | 公開日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.601 Å) | | 主引用文献 | Crystal structure of AtHPPD-Y19788 complex
To Be Published
|
|
2YMX
 
 | | Crystal structure of inhibitory anti-AChE Fab408 | | 分子名称: | FAB ANTIBODY HEAVY CHAIN, FAB ANTIBODY LIGHT CHAIN, GLYCEROL | | 著者 | Bourne, Y, Renault, L, Essono, S, Mondielli, G, Lamourette, P, Bocquet, D, Grassi, J, Marchot, P. | | 登録日 | 2012-10-10 | | 公開日 | 2013-10-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular Characterization of Monoclonal Antibodies that Inhibit Acetylcholinesterase by Targeting the Peripheral Site and Backdoor Region
Plos One, 8, 2013
|
|
1GFE
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-12-04 | | 公開日 | 2000-12-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
6B7P
 
 | |
1G0G
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152A | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | 著者 | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | 登録日 | 2000-10-06 | | 公開日 | 2001-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
7ZRG
 
 | | Cryo-EM map of the WT KdpFABC complex in the E1_ATPearly conformation, under turnover conditions | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | 著者 | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Rheinberger, J, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | 登録日 | 2022-05-04 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
3K21
 
 | | Crystal Structure of carboxy-terminus of PFC0420w. | | 分子名称: | ACETATE ION, CALCIUM ION, Calcium-dependent protein kinase 3, ... | | 著者 | Wernimont, A.K, Hutchinson, A, Artz, J.D, Mackenzie, F, Cossar, D, Kozieradzki, I, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | 登録日 | 2009-09-29 | | 公開日 | 2010-01-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Structures of parasitic CDPK domains point to a common mechanism of activation.
Proteins, 79, 2011
|
|
1G0K
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152C | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | 著者 | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | 登録日 | 2000-10-06 | | 公開日 | 2001-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
5AHE
 
 | | Crystal structure of Salmonella enterica HisA | | 分子名称: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | 著者 | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | 登録日 | 2015-02-05 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Two-Step Ligand Binding in a Beta/Alpha8 Barrel Enzyme -Substrate-Bound Structures Shed New Light on the Catalytic Cycle of Hisa
J.Biol.Chem., 290, 2015
|
|
8JKR
 
 | | SP1746 in complex with UMP | | 分子名称: | 1,2-ETHANEDIOL, FE (III) ION, GLYCEROL, ... | | 著者 | Jin, Y, Niu, L, Ke, J. | | 登録日 | 2023-06-01 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 2024
|
|
4PC9
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM ROSENBACTER DENITRIFICANS OCh 114 (RD1_1052, TARGET EFI-510238) WITH BOUND D-MANNONATE | | 分子名称: | C4-dicarboxylate transport system, substrate-binding protein, putative, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-04-14 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
5JNP
 
 | | Crystal structure of a rice (Oryza Sativa) cellulose synthase plant conserved region (P-CR) | | 分子名称: | CITRATE ANION, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Rushton, P.S, Olek, A.T, Makowski, L, Badger, J, Steussy, C.N, Carpita, N.C, Stauffacher, C.V. | | 登録日 | 2016-04-30 | | 公開日 | 2016-12-28 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.404 Å) | | 主引用文献 | Rice Cellulose SynthaseA8 Plant-Conserved Region Is a Coiled-Coil at the Catalytic Core Entrance.
Plant Physiol., 173, 2017
|
|
8A92
 
 | | p53-Y220C Core Domain in Complex with a Bromo-trifluoro-pyrazole-amine | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-bromanyl-5-(trifluoromethyl)-1H-pyrazol-3-amine, Cellular tumor antigen p53, ... | | 著者 | Stahlecker, J, Braun, M.B, Stehle, T, Boeckler, F.M. | | 登録日 | 2022-06-27 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Revisiting a challenging p53 binding site: a diversity-optimized HEFLib reveals diverse binding modes in T-p53C-Y220C.
Rsc Med Chem, 13, 2022
|
|
8JK5
 
 | |
2YNT
 
 | | GIM-1-3Mol native. Crystal structures of Pseudomonas aeruginosa GIM- 1: active site plasticity in metallo-beta-lactamases | | 分子名称: | GIM-1 PROTEIN, GLYCEROL, ZINC ION | | 著者 | Borra, P.S, Samuelsen, O, Spencer, J, Lorentzen, M.S, Leiros, H.-K.S. | | 登録日 | 2012-10-18 | | 公開日 | 2013-07-24 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.598 Å) | | 主引用文献 | Crystal Structures of Pseudomonas Aeruginosa Gim-1: Active-Site Plasticity in Metallo-Beta-Lactamases.
Antimicrob.Agents Chemother., 57, 2013
|
|
1SH2
 
 | | Crystal Structure of Norwalk Virus Polymerase (Metal-free, Centered Orthorhombic) | | 分子名称: | RNA Polymerase | | 著者 | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | 登録日 | 2004-02-24 | | 公開日 | 2004-03-09 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
8JK9
 
 | | SP1746 in complex with GDP | | 分子名称: | FE (III) ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Jin, Y, Ke, J, Niu, L. | | 登録日 | 2023-06-01 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 2024
|
|
8J8X
 
 | | Crystal structure of AtHPPD-YH20307 complex | | 分子名称: | 4-hydroxyphenylpyruvate dioxygenase, 5-(1,3-dimethyl-5-oxidanyl-pyrazol-4-yl)carbonyl-1,4-dimethyl-3-propan-2-yl-benzimidazol-2-one, COBALT (II) ION | | 著者 | Lin, H.-Y, Dong, J, Yang, G.-F. | | 登録日 | 2023-05-02 | | 公開日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.598 Å) | | 主引用文献 | Crystal structure of AtHPPD-YH20307 complex
To Be Published
|
|
