7WPN
 
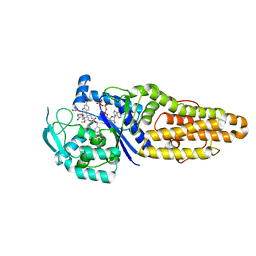 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a phenylbenzimidazole inhibitor and ATP | | 分子名称: | (phenylmethyl) N-[(2R)-2-[2-(4-bromanyl-3-oxidanyl-phenyl)-5-(methylcarbamoyl)benzimidazol-1-yl]-2-(3,4-dimethoxyphenyl)ethyl]carbamate, 1,2-ETHANEDIOL, ACETIC ACID, ... | | 著者 | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | 登録日 | 2022-01-24 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPL
 
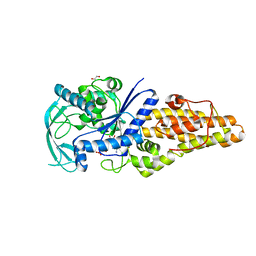 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with ATP | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Methionine--tRNA ligase | | 著者 | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | 登録日 | 2022-01-24 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
6HUK
 
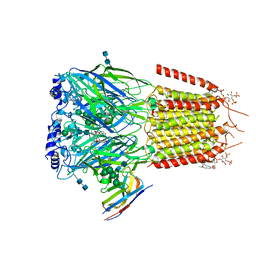 | | CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with bicuculline and megabody Mb38. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-1,Gamma-aminobutyric acid receptor subunit alpha-1, Gamma-aminobutyric acid receptor subunit beta-3, ... | | 著者 | Masiulis, S, Desai, R, Uchanski, T, Serna Martin, I, Laverty, D, Karia, D, Malinauskas, T, Jasenko, Z, Pardon, E, Kotecha, A, Steyaert, J, Miller, K.W, Aricescu, A.R. | | 登録日 | 2018-10-08 | | 公開日 | 2019-01-02 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.69 Å) | | 主引用文献 | GABAAreceptor signalling mechanisms revealed by structural pharmacology.
Nature, 565, 2019
|
|
5WQT
 
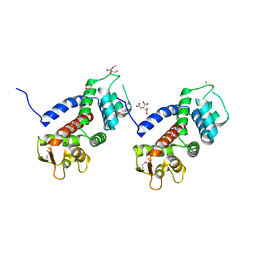 | | Structure of a protein involved in pyroptosis | | 分子名称: | CITRIC ACID, GLYCEROL, Gasdermin-D | | 著者 | Kuang, S, Li, J. | | 登録日 | 2016-11-28 | | 公開日 | 2017-10-04 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Structure insight of GSDMD reveals the basis of GSDMD autoinhibition in cell pyroptosis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7WPJ
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus | | 分子名称: | 1,2-ETHANEDIOL, Methionine--tRNA ligase | | 著者 | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | 登録日 | 2022-01-24 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
7WPI
 
 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a phenylbenzimidazole inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-(4-bromanyl-3-oxidanyl-phenyl)-1-[(1R)-2-[2-(2-tert-butylphenoxy)ethanoylamino]-1-(3,4-dimethoxyphenyl)ethyl]-N-methyl-benzimidazole-5-carboxamide, ACETATE ION, ... | | 著者 | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | 登録日 | 2022-01-24 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
3MV5
 
 | | Crystal structure of Akt-1-inhibitor complexes | | 分子名称: | (3R)-1-(5-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-3-amine, GSK3-beta peptide, MANGANESE (II) ION, ... | | 著者 | Pandit, J. | | 登録日 | 2010-05-03 | | 公開日 | 2010-06-02 | | 最終更新日 | 2021-10-06 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Design of selective, ATP-competitive inhibitors of Akt.
J.Med.Chem., 53, 2010
|
|
3WUF
 
 | | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 | | 分子名称: | Endo-1,4-beta-xylanase A, ZINC ION | | 著者 | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | 登録日 | 2014-04-23 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
4QZA
 
 | | Mouse Tdt in complex with a DSB substrate, C-C base pair | | 分子名称: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*TP*TP*TP*TP*GP*C)-3', ... | | 著者 | Gouge, J, Delarue, M. | | 登録日 | 2014-07-27 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair.
Embo J., 34, 2015
|
|
4QZD
 
 | | Mouse Tdt, F405A mutant, in complex with a DSB substrate, C-C base pair | | 分子名称: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*TP*TP*TP*TP*GP*C)-3', ... | | 著者 | Gouge, J, Delarue, M. | | 登録日 | 2014-07-27 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair.
Embo J., 34, 2015
|
|
3MVJ
 
 | | Human cyclic AMP-dependent protein kinase PKA inhibitor complex | | 分子名称: | (3R)-1-(5-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)pyrrolidin-3-amine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | 著者 | Pandit, J, Vajdos, F. | | 登録日 | 2010-05-04 | | 公開日 | 2010-06-02 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Design of selective, ATP-competitive inhibitors of Akt.
J.Med.Chem., 53, 2010
|
|
4QZH
 
 | | Mouse Tdt, F401A mutant, in complex with a DSB substrate, C-T base pair | | 分子名称: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*TP*TP*TP*TP*GP*T)-3', ... | | 著者 | Gouge, J, Delarue, M. | | 登録日 | 2014-07-27 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair.
Embo J., 34, 2015
|
|
5YOV
 
 | | Crystal structure of BRD4-BD1 bound with hjp126 | | 分子名称: | (3~{R})-4-cyclopentyl-~{N}-(2,4-dimethylphenyl)-1,3-dimethyl-2-oxidanylidene-3~{H}-quinoxaline-6-carboxamide, Bromodomain-containing protein 4 | | 著者 | Xiong, B, Hu, J, Li, Y, Cao, D. | | 登録日 | 2017-10-31 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.451 Å) | | 主引用文献 | Structure-based optimization of a series of selective BET inhibitors containing aniline or indoline groups.
Eur.J.Med.Chem., 150, 2018
|
|
2NXW
 
 | | Crystal structure of phenylpyruvate decarboxylase of Azospirillum brasilense | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Versees, W, Spaepen, S, Vanderleyden, J, Steyaert, J. | | 登録日 | 2006-11-20 | | 公開日 | 2007-05-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The crystal structure of phenylpyruvate decarboxylase from Azospirillum brasilense at 1.5 A resolution. Implications for its catalytic and regulatory mechanism.
Febs J., 274, 2007
|
|
6HJX
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) 7'C pore mutant (L238C) in complex with nanobody 72 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cys-loop ligand-gated ion channel, DODECYL-BETA-D-MALTOSIDE, ... | | 著者 | Spurny, R, Govaerts, C, Evans, G.L, Pardon, E, Steyaert, J, Ulens, C. | | 登録日 | 2018-09-04 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A lipid site shapes the agonist response of a pentameric ligand-gated ion channel.
Nat.Chem.Biol., 15, 2019
|
|
6HLK
 
 | | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit. | | 分子名称: | Redirecting phage packaging protein C (RppC) | | 著者 | Penades, J.R, Bacarizo, J, Marina, A, Alqasmi, M, Fillol-Salom, A, Roszak, A.W, Ciges-Tomas, J.R. | | 登録日 | 2018-09-11 | | 公開日 | 2019-07-31 | | 最終更新日 | 2019-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit.
Mol.Cell, 75, 2019
|
|
7Q3O
 
 | |
7E35
 
 | | Crystal structure of the SARS-CoV-2 papain-like protease (PLPro) C112S mutant bound to compound S43 | | 分子名称: | N-[(3-acetamidophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Non-structural protein 3, ZINC ION | | 著者 | Liu, J, Wang, Y, Xu, X, Pan, L. | | 登録日 | 2021-02-08 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2.
Cell Chem Biol, 28, 2021
|
|
7PJH
 
 | | Crystal structure of the human spliceosomal maturation factor AAR2 bound to the RNAse H domain of PRPF8 | | 分子名称: | Pre-mRNA-processing-splicing factor 8, Protein AAR2 homolog | | 著者 | Preussner, M, Santos, K, Heroven, A.C, Alles, J, Heyd, F, Wahl, M.C, Weber, G. | | 登録日 | 2021-08-24 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural and functional investigation of the human snRNP assembly factor AAR2 in complex with the RNase H-like domain of PRPF8.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3KG1
 
 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, mutant N63A | | 分子名称: | CHLORIDE ION, SnoaB | | 著者 | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | 登録日 | 2009-10-28 | | 公開日 | 2010-01-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
6BCO
 
 | | cryo-EM structure of TRPM4 in ATP bound state with short coiled coil at 2.9 angstrom resolution | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Transient receptor potential cation channel subfamily M member 4 | | 著者 | Guo, J, She, J, Chen, Q, Bai, X, Jiang, Y. | | 登録日 | 2017-10-20 | | 公開日 | 2017-12-13 | | 最終更新日 | 2019-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.88 Å) | | 主引用文献 | Structures of the calcium-activated, non-selective cation channel TRPM4.
Nature, 552, 2017
|
|
7Q4N
 
 | |
2J6Y
 
 | | Structural and Functional Characterisation of partner switching regulating the environmental stress response in Bacillus subtilis | | 分子名称: | PHOSPHOSERINE PHOSPHATASE RSBU | | 著者 | Hardwick, S.W, Pane-Farre, J, Delumeau, O, Marles-Wright, J, Murray, J.W, Hecker, M, Lewis, R.J. | | 登録日 | 2006-10-05 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and functional characterization of partner switching regulating the environmental stress response in Bacillus subtilis.
J. Biol. Chem., 282, 2007
|
|
6IZI
 
 | |
6J0A
 
 | |
