5CMV
 
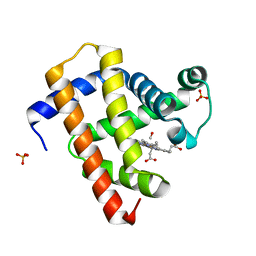 | | Ultrafast dynamics in myoglobin: dark-state, CO-ligated structure | | 分子名称: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
5CND
 
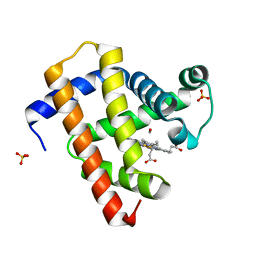 | | Ultrafast dynamics in myoglobin: 3 ps time delay | | 分子名称: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
5J0L
 
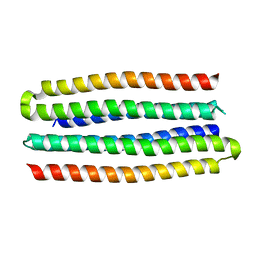 | | De novo design of protein homo-oligomers with modular hydrogen bond network-mediated specificity | | 分子名称: | designed protein 3L6HC2_2 | | 著者 | Sankaran, B, Zwart, P.H, Pereira, J.H, Baker, D, Boyken, S, Chen, Z, Groves, B, Langan, R.A, Oberdorfer, G, Ford, A, Gilmore, J, Xu, C, DiMaio, F, Seelig, G. | | 登録日 | 2016-03-28 | | 公開日 | 2016-07-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity.
Science, 352, 2016
|
|
8PN8
 
 | | Engineered glycolyl-CoA carboxylase (L100N variant) with bound CoA | | 分子名称: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | 著者 | Zarzycki, J, Marchal, D.G, Schulz, L, Prinz, S, Erb, T.J. | | 登録日 | 2023-06-30 | | 公開日 | 2023-11-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (2.31 Å) | | 主引用文献 | Machine Learning-Supported Enzyme Engineering toward Improved CO 2 -Fixation of Glycolyl-CoA Carboxylase.
Acs Synth Biol, 12, 2023
|
|
4QS9
 
 | |
3JPN
 
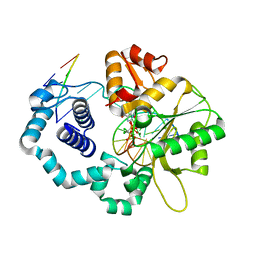 | | Ternary complex of DNA polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-dichloro methylene triphosphate | | 分子名称: | 2'-deoxy-5'-O-[(S)-{[(R)-[dichloro(phosphono)methyl](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]guanosine, 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3', ... | | 著者 | Batra, V.K, Upton, J, Kashmerov, B, Beard, W.A, Wilson, S.H, Goodman, M.F, McKenna, C.E. | | 登録日 | 2009-09-04 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Halogenated beta,gamma-Methylene- and Ethylidene-dGTP-DNA Ternary Complexes with DNA Polymerase beta: Structural Evidence for Stereospecific Binding of the Fluoromethylene Analogues.
J.Am.Chem.Soc., 132, 2010
|
|
3JPR
 
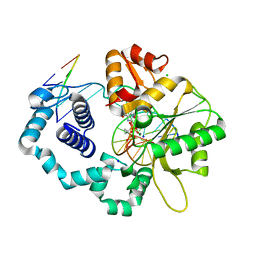 | | Ternary complex of DNA polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-dimethyl methylene triphosphate | | 分子名称: | 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(1-methyl-1-phosphonoethyl)phosphoryl]oxy}phosphoryl]guanosine, 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DOC))-3', ... | | 著者 | Batra, V.K, Upton, J, Kashmerov, B, Beard, W.A, Wilson, S.H, Goodman, M.F, McKenna, C.E. | | 登録日 | 2009-09-04 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Halogenated beta,gamma-Methylene- and Ethylidene-dGTP-DNA Ternary Complexes with DNA Polymerase beta: Structural Evidence for Stereospecific Binding of the Fluoromethylene Analogues.
J.Am.Chem.Soc., 132, 2010
|
|
6N4Q
 
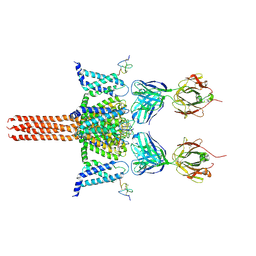 | | CryoEM structure of Nav1.7 VSD2 (actived state) in complex with the gating modifier toxin ProTx2 | | 分子名称: | Beta/omega-theraphotoxin-Tp2a, Fab heavy chain, Fab light chain, ... | | 著者 | Xu, H, Rohou, A, Arthur, C.P, Estevez, A, Ciferri, C, Payandeh, J, Koth, C.M. | | 登録日 | 2018-11-20 | | 公開日 | 2019-01-23 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin.
Cell, 176, 2019
|
|
4QZ8
 
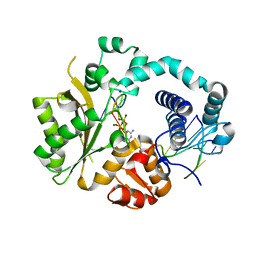 | | Mouse Tdt in complex with a DSB substrate, C-G base pair | | 分子名称: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*TP*TP*TP*TP*GP*G)-3', ... | | 著者 | Gouge, J, Delarue, M. | | 登録日 | 2014-07-27 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair.
Embo J., 34, 2015
|
|
4QZC
 
 | | Mouse Tdt, F405A mutant, in complex with a DSB substrate, C-G base pair | | 分子名称: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*TP*TP*TP*TP*GP*G)-3', ... | | 著者 | Gouge, J, Delarue, M. | | 登録日 | 2014-07-27 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair.
Embo J., 34, 2015
|
|
4QZG
 
 | | Mouse Tdt, F401A mutant, in complex with a DSB substrate, C-C base pair | | 分子名称: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*TP*TP*TP*TP*GP*C)-3', ... | | 著者 | Gouge, J, Delarue, M. | | 登録日 | 2014-07-27 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair.
Embo J., 34, 2015
|
|
5J99
 
 | | Ambient temperature transition state structure of arginine kinase - crystal 8/Form I | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | 著者 | Godsey, M, Davulcu, O, Nix, J, Skalicky, J.J, Bruschweiler, R, Chapman, M.S. | | 登録日 | 2016-04-08 | | 公開日 | 2016-08-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Sampling of Conformational Dynamics in Ambient-Temperature Crystal Structures of Arginine Kinase.
Structure, 24, 2016
|
|
7SW4
 
 | | MicroED structure of proteinase K from a 540 nm thick lamella measured at 200 kV | | 分子名称: | Proteinase K | | 著者 | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | 登録日 | 2021-11-19 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.4 Å) | | 主引用文献 | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW9
 
 | | MicroED structure of proteinase K from a 170 nm thick lamella measured at 300 kV | | 分子名称: | Proteinase K | | 著者 | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | 登録日 | 2021-11-19 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.1 Å) | | 主引用文献 | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SVY
 
 | | MicroED structure of proteinase K from a 130 nm thick lamella measured at 120 kV | | 分子名称: | Proteinase K | | 著者 | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | 登録日 | 2021-11-19 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.3 Å) | | 主引用文献 | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
197L
 
 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | 著者 | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | 登録日 | 1995-11-06 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
7SWB
 
 | | MicroED structure of proteinase K from a 360 nm thick lamella measured at 300 kV | | 分子名称: | Proteinase K | | 著者 | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | 登録日 | 2021-11-19 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.05 Å) | | 主引用文献 | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW2
 
 | | MicroED structure of proteinase K from a 130 nm thick lamella measured at 200 kV | | 分子名称: | Proteinase K | | 著者 | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | 登録日 | 2021-11-19 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.95 Å) | | 主引用文献 | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW8
 
 | | MicroED structure of proteinase K from a 150 nm thick lamella measured at 300 kV | | 分子名称: | Proteinase K | | 著者 | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | 登録日 | 2021-11-19 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | 主引用文献 | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5JJJ
 
 | | Structure of the SRII/HtrII Complex in P64 space group ("U" shape) | | 分子名称: | EICOSANE, RETINAL, Sensory rhodopsin II transducer, ... | | 著者 | Ishchenko, A, Round, E, Borshchevskiy, V, Grudinin, S, Gushchin, I, Klare, J, Remeeva, A, Polovinkin, V, Utrobin, P, Balandin, T, Engelhard, M, Bueldt, G, Gordeliy, V. | | 登録日 | 2016-04-24 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | New Insights on Signal Propagation by Sensory Rhodopsin II/Transducer Complex.
Sci Rep, 7, 2017
|
|
5CN8
 
 | | Ultrafast dynamics in myoglobin: 0.3 ps time delay | | 分子名称: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
5CNF
 
 | | Ultrafast dynamics in myoglobin: 50 ps time delay | | 分子名称: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | 登録日 | 2015-07-17 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
6N4I
 
 | |
7SW5
 
 | | MicroED structure of proteinase K from a 460 nm thick lamella measured at 200 kV | | 分子名称: | Proteinase K | | 著者 | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | 登録日 | 2021-11-19 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.95 Å) | | 主引用文献 | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4R8U
 
 | | S-SAD structure of DINB-DNA Complex | | 分子名称: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA, DNA polymerase IV, ... | | 著者 | Kottur, J, Nair, D.T, Weinert, T, Oligeric, V, Wang, M. | | 登録日 | 2014-09-03 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Fast native-SAD phasing for routine macromolecular structure determination
Nat.Methods, 12, 2015
|
|
