1NXQ
 
 | | Crystal Structure of R-alcohol dehydrogenase (RADH) (apoenyzme) from Lactobacillus brevis | | 分子名称: | MAGNESIUM ION, R-alcohol dehydrogenase | | 著者 | Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | 登録日 | 2003-02-11 | | 公開日 | 2003-04-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | The crystal structure of R-specific alcohol dehydrogenase from Lactobacillus brevis suggests the structural basis of its metal dependency
J.Mol.Biol., 327, 2003
|
|
3ZD8
 
 | | Potassium bound structure of E. coli ExoIX in P1 | | 分子名称: | POTASSIUM ION, PROTEIN XNI | | 著者 | Anstey-Gilbert, C.S, Hemsworth, G.R, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | 登録日 | 2012-11-26 | | 公開日 | 2013-07-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
3JAC
 
 | | Cryo-EM study of a channel | | 分子名称: | Piezo-type mechanosensitive ion channel component 1 | | 著者 | Ge, J, Li, W, Zhao, Q, Li, N, Xiao, B, Gao, N, Yang, M. | | 登録日 | 2015-06-05 | | 公開日 | 2015-09-23 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Architecture of the mammalian mechanosensitive Piezo1 channel
Nature, 527, 2015
|
|
3JSZ
 
 | | Legionella pneumophila glucosyltransferase Lgt1 N293A with UDP-Glc | | 分子名称: | MAGNESIUM ION, Putative uncharacterized protein, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | 著者 | Lu, W, Du, J, Belyi, Y, Stahl, M, Zivilikidis, T, Gerhardt, S, Aktories, K, Einsle, O. | | 登録日 | 2009-09-11 | | 公開日 | 2010-02-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Basis of the Action of Glucosyltransferase Lgt1 from Legionella pneumophila.
J.Mol.Biol., 2009
|
|
3ZKR
 
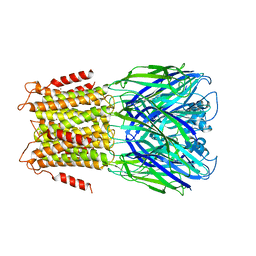 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with bromoform | | 分子名称: | CYS-LOOP LIGAND-GATED ION CHANNEL, TRIBROMOMETHANE | | 著者 | Spurny, R, Billen, B, Howard, R.J, Brams, M, Debaveye, S, Price, K.L, Weston, D.A, Strelkov, S.V, Tytgat, J, Bertrand, S, Bertrand, D, Lummis, S.C.R, Ulens, C. | | 登録日 | 2013-01-24 | | 公開日 | 2013-02-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.649 Å) | | 主引用文献 | Multisite Binding of a General Anesthetic to the Prokaryotic Pentameric Erwinia Chrysanthemi Ligand-Gated Ion Channel (Elic).
J.Biol.Chem., 288, 2013
|
|
3JTN
 
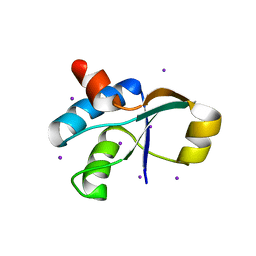 | | Crystal Structure of the c-terminal domain of YpbH | | 分子名称: | Adapter protein mecA 2, IODIDE ION | | 著者 | Wang, F, Mei, Z, Qi, Y, Yan, C, Wang, J, Shi, Y. | | 登録日 | 2009-09-14 | | 公開日 | 2009-09-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Crystal Structure of the MecA degradation tag
To be Published
|
|
3ZR8
 
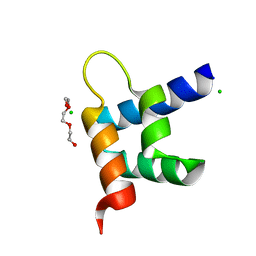 | | Crystal structure of RxLR effector Avr3a11 from Phytophthora capsici | | 分子名称: | AVR3A11, CHLORIDE ION, TRIETHYLENE GLYCOL | | 著者 | Boutemy, L.S, King, S.R.F, Win, J, Hughes, R.K, Clarke, T.A, Blumenschein, T.M.A, Kamoun, S, Banfield, M.J. | | 登録日 | 2011-06-15 | | 公開日 | 2011-08-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Structures of Phytophthora Rxlr Effector Proteins: A Conserved But Adaptable Fold Underpins Functional Diversity.
J.Biol.Chem., 286, 2011
|
|
3JTP
 
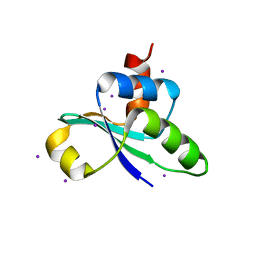 | | crystal structure of the C-terminal domain of MecA | | 分子名称: | Adapter protein mecA 1, IODIDE ION | | 著者 | Wang, F, Mei, Z, Qi, Y, Yan, C, Wang, J, Shi, Y. | | 登録日 | 2009-09-14 | | 公開日 | 2009-09-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | crystal structure of the MecA degradation tag
To be Published
|
|
3JV2
 
 | | Crystal Structure of B. subtilis SecA with bound peptide | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein translocase subunit SecA, ... | | 著者 | Zimmer, J. | | 登録日 | 2009-09-15 | | 公開日 | 2009-11-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Conformational flexibility and peptide interaction of the translocation ATPase SecA.
J.Mol.Biol., 394, 2009
|
|
3JW8
 
 | | Crystal structure of human mono-glyceride lipase | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, MGLL protein | | 著者 | Bertrand, T, Auge, F, Houtmann, J, Rak, A, Vallee, F, Mikol, V, Berne, P.F, Michot, N, Cheuret, D, Hoornaert, C, Mathieu, M. | | 登録日 | 2009-09-18 | | 公開日 | 2010-01-26 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for human monoglyceride lipase inhibition.
J.Mol.Biol., 396, 2010
|
|
3JXQ
 
 | | X-Ray structure of r[CGCG(5-fluoro)CG]2 | | 分子名称: | MAGNESIUM ION, r[CGCG(5-fluoro)CG]2 | | 著者 | Adamiak, D.A, Milecki, J, Adamiak, R.W, Rypniewski, W. | | 登録日 | 2009-09-21 | | 公開日 | 2010-02-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The hydration and unusual hydrogen bonding in the crystal structure of an RNA duplex containing alternating CG base pairs
New J.Chem., 34, 2010
|
|
3ZPT
 
 | | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors | | 分子名称: | CHLORIDE ION, PROTEASE, methyl N-[(2S)-1-[2-[(4-bromophenyl)methyl]-2-[3-[(3Z,8S,11R)-11-oxidanyl-7,10-bis(oxidanylidene)-8-propan-2-yl-6,9-diazabicyclo[11.2.2]heptadeca-1(16),3,13(17),14-tetraen-11-yl]propyl]hydrazinyl]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | 著者 | Joshi, A, Veron, J.B, Unge, J, Rosenquist, A, Wallberg, H, Samuelsson, B, Hallberg, A, Larhed, M. | | 登録日 | 2013-03-01 | | 公開日 | 2013-11-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Design and Synthesis of P1-P3 Macrocyclic Tertiary Alcohol Comprising HIV-1 Protease Inhibitors
J.Med.Chem., 56, 2013
|
|
3JB8
 
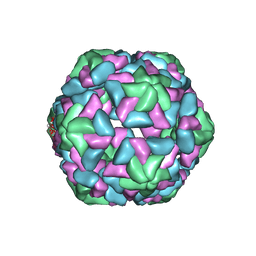 | | Insight into Three-dimensional structure of Maize Chlorotic Mottle Virus Revealed by Single Particle Analysis | | 分子名称: | Coat protein | | 著者 | Wang, C.Y, Zhang, Q.F, Gao, Y.Z, Zhou, X.P, Ji, G, Huang, X.J, Hong, J, Zhang, C.X. | | 登録日 | 2015-08-04 | | 公開日 | 2016-07-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Insight into the three-dimensional structure of maize chlorotic mottle virus revealed by Cryo-EM single particle analysis.
Virology, 485, 2015
|
|
4AA8
 
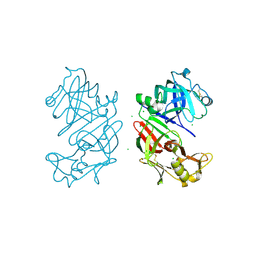 | | Bovine chymosin at 1.8A resolution | | 分子名称: | CHLORIDE ION, CHYMOSIN | | 著者 | Langholm Jensen, J, Molgaard, A, Navarro Poulsen, J.C, van den Brink, J.M, Harboe, M, Simonsen, J.B, Qvist, K.B, Larsen, S. | | 登録日 | 2011-11-30 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Camel and Bovine Chymosin: The Relationship between Their Structures and Cheese-Making Properties.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4A5S
 
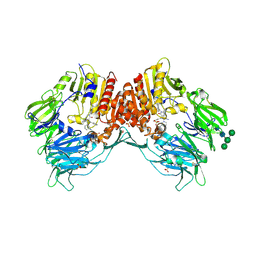 | | CRYSTAL STRUCTURE OF HUMAN DPP4 IN COMPLEX WITH A NOVAL HETEROCYCLIC DPP4 INHIBITOR | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[(3S)-3-AMINOPIPERIDIN-1-YL]-5-BENZYL-4-OXO-3-(QUINOLIN-4-YLMETHYL)-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDINE-7-CARBONITRILE, DIPEPTIDYL PEPTIDASE 4 SOLUBLE FORM, ... | | 著者 | Ostermann, N, Kroemer, M, Zink, F, Gerhartz, B, Sutton, J.M, Clark, D.E, Dunsdon, S.J, Fenton, G, Fillmore, A, Harris, N.V, Higgs, C, Hurley, C.A, Krintel, S.L, MacKenzie, R.E, Duttaroy, A, Gangl, E, Maniara, W, Sedrani, R, Namoto, K, Sirockin, F, Trappe, J, Hassiepen, U, Baeschlin, D.K. | | 登録日 | 2011-10-28 | | 公開日 | 2012-02-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Novel Heterocyclic Dpp-4 Inhibitors for the Treatment of Type 2 Diabetes.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3K18
 
 | |
1RNA
 
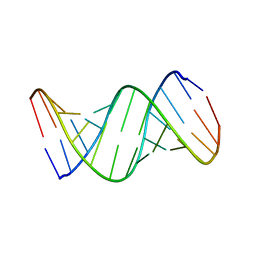 | | CRYSTALLOGRAPHIC STRUCTURE OF AN RNA HELIX: [U(U-A)6A]2 | | 分子名称: | RNA (5'-R(*UP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*A)-3') | | 著者 | Dock-Bregeon, A.C, Chevrier, B, Podjarny, A, Johnson, J, De Bear, J.S, Gough, G.R, Gilham, P.T, Moras, D. | | 登録日 | 1990-02-01 | | 公開日 | 1991-04-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystallographic structure of an RNA helix: [U(UA)6A]2.
J.Mol.Biol., 209, 1989
|
|
4AAQ
 
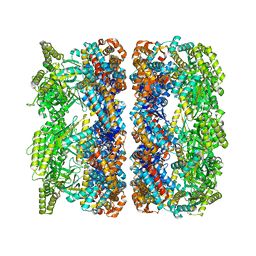 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | 分子名称: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | 登録日 | 2011-12-05 | | 公開日 | 2012-12-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
4A9Z
 
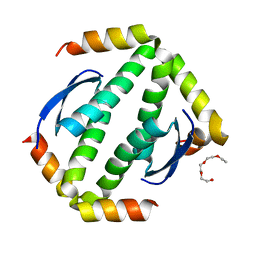 | | CRYSTAL STRUCTURE OF HUMAN P63 TETRAMERIZATION DOMAIN | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, TUMOR PROTEIN 63 | | 著者 | Muniz, J.R.C, Coutandin, D, Salah, E, Chaikuad, A, Vollmar, M, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, von Delft, F, Knapp, S. | | 登録日 | 2011-11-30 | | 公開日 | 2011-12-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Crystal Structure of Human P63 Tetramerization Domain
To be Published
|
|
3J8W
 
 | |
3J9C
 
 | | CryoEM single particle reconstruction of anthrax toxin protective antigen pore at 2.9 Angstrom resolution | | 分子名称: | CALCIUM ION, Protective antigen PA-63 | | 著者 | Jiang, J, Pentelute, B.L, Collier, R.J, Zhou, Z.H. | | 登録日 | 2014-12-25 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Atomic structure of anthrax protective antigen pore elucidates toxin translocation.
Nature, 521, 2015
|
|
3JQJ
 
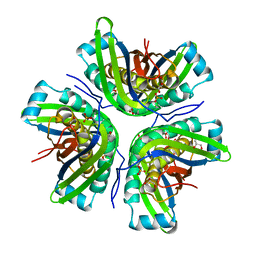 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 | | 分子名称: | GLYCEROL, Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION, ... | | 著者 | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2009-09-07 | | 公開日 | 2010-06-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3J9U
 
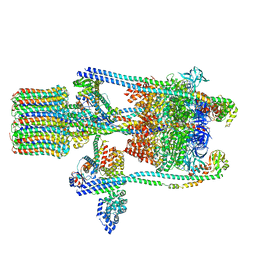 | | Yeast V-ATPase state 2 | | 分子名称: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | 著者 | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | 登録日 | 2015-02-23 | | 公開日 | 2015-05-13 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
4AMM
 
 | |
3JR2
 
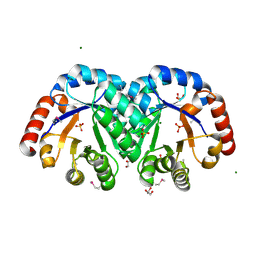 | | X-ray crystal structure of the Mg-bound 3-keto-L-gulonate-6-phosphate decarboxylase from Vibrio cholerae O1 biovar El Tor str. N16961 | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Hexulose-6-phosphate synthase SgbH, ... | | 著者 | Nocek, B, Maltseva, N, Stam, J, Anderson, W, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-09-08 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the Mg-bound 3-keto-L-gulonate-6-phosphate
decarboxylase from Vibrio cholerae O1 biovar El Tor str. N16961
To be Published
|
|
