8J60
 
 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 protein, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
8J21
 
 | | Cryo-EM structure of FFAR3 complex bound with butyrate acid | | Descriptor: | Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
5EN2
 
 | | Molecular basis for antibody-mediated neutralization of New World hemorrhagic fever mammarenaviruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Mahmutovic, S, Clark, L, Levis, S, Briggiler, A, Enria, D, Harrison, S.C, Abraham, J. | | Deposit date: | 2015-11-09 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Molecular Basis for Antibody-Mediated Neutralization of New World Hemorrhagic Fever Mammarenaviruses.
Cell Host Microbe, 18, 2015
|
|
5EOA
 
 | | Crystal structure of OPTN E50K mutant and TBK1 complex | | Descriptor: | Optineurin, Serine/threonine-protein kinase TBK1 | | Authors: | Li, F, Xie, X, Liu, J, Pan, L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
1YPL
 
 | |
8J5Y
 
 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 isoform 1, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
1YPX
 
 | | Crystal Structure of the Putative Vitamin-B12 Independent Methionine Synthase from Listeria monocytogenes, Northeast Structural Genomics Target LmR13 | | Descriptor: | PHOSPHATE ION, putative vitamin-B12 independent methionine synthase family protein | | Authors: | Forouhar, F, Chen, Y, Benach, J, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-01-31 | | Release date: | 2005-02-08 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Putative Vitamin-B12 Independent Methionine Synthase from Listeria monocytogenes, Northeast Structural Genomics Target LmR13
To be Published
|
|
4QQ3
 
 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP | | Descriptor: | CHLORIDE ION, Inosine-5'-monophosphate dehydrogenase, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-26 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from vibrio cholerae, deletion mutant, in complex with xmp
To be Published
|
|
5EQ0
 
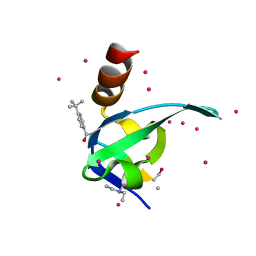 | | Crystal Structure of chromodomain of CBX8 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 8, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-12 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
8JM9
 
 | |
4R70
 
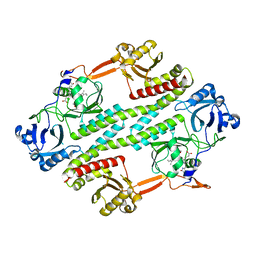 | |
8JMI
 
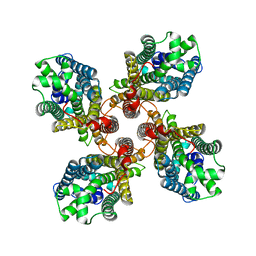 | |
1YR5
 
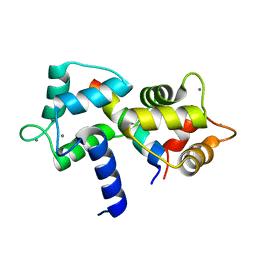 | |
5EGG
 
 | |
1YSN
 
 | | Solution structure of the anti-apoptotic protein Bcl-xL complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-X | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1W90
 
 | | CBM29-2 mutant D114A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | 1,2-ETHANEDIOL, NON-CATALYTIC PROTEIN 1, SODIUM ION | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-10-01 | | Release date: | 2005-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
5EFZ
 
 | | Monoclinic structure of the acetyl esterase MekB | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Toelzer, C, Pal, S, Watzlawick, H, Altenbuchner, J, Niefind, K. | | Deposit date: | 2015-10-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A novel esterase subfamily with alpha / beta-hydrolase fold suggested by structures of two bacterial enzymes homologous to l-homoserine O-acetyl transferases.
Febs Lett., 590, 2016
|
|
8JKE
 
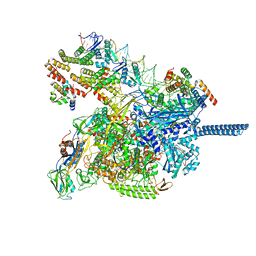 | | AfsR(T337A) transcription activation complex | | Descriptor: | DNA(65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wang, Y, Zheng, J. | | Deposit date: | 2023-06-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural and functional characterization of AfsR, an SARP family transcriptional activator of antibiotic biosynthesis in Streptomyces.
Plos Biol., 22, 2024
|
|
1W8Z
 
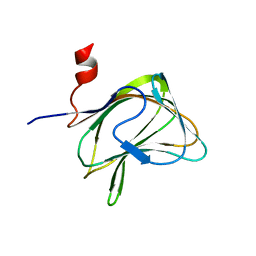 | | CBM29-2 mutant K85A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1 | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-10-01 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
8JMA
 
 | |
8JME
 
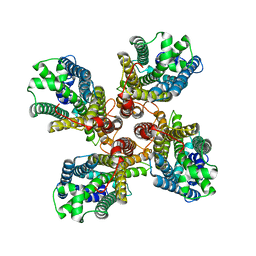 | |
8JMH
 
 | |
1W8B
 
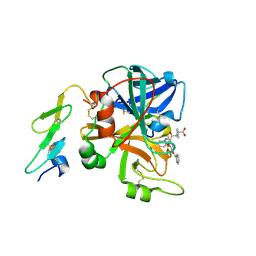 | | Factor7 - 413 complex | | Descriptor: | (S)-[(R)-2-(4-BENZYLOXY-3-METHOXY-PHENYL)-2-(4-CARBAMIMIDOYL-PHENYLAMINO)-ACETYLAMINO]-PHENYL-ACETIC ACID, BLOOD COAGULATION FACTOR VIIA, CALCIUM ION | | Authors: | Ackermann, J, Alig, L, Banner, D.W, Boehm, H.-J, Groebke-Zbinden, K, Hilpert, K, Lave, T, Kuehne, H, Obst-Sander, U, Riederer, M.A, Stahl, M, Tschopp, T.B, Weber, L, Wessel, H.P. | | Deposit date: | 2004-09-17 | | Release date: | 2005-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Selective and Orally Bioavailable Phenylglycine Tissue Factor/Factor Viia Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1W63
 
 | | AP1 clathrin adaptor core | | Descriptor: | ADAPTER-RELATED PROTEIN COMPLEX 1 BETA 1 SUBUNIT, ADAPTER-RELATED PROTEIN COMPLEX 1 GAMMA 1 SUBUNIT, ADAPTER-RELATED PROTEIN COMPLEX 1 SIGMA 1A SUBUNIT, ... | | Authors: | Heldwein, E, Macia, E, Wang, J, Yin, H.L, Kirchhausen, T, Harrison, S.C. | | Deposit date: | 2004-08-12 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of the Clathrin Adaptor Protein 1 Core
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5DL9
 
 | | Structure of Tetragonal Lysozyme in complex with Iodine solved by UWO Students | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IODIDE ION, ... | | Authors: | Bednarski, R, Cirricione, N, Greco, A, Hodgson, R, Kent, S, McGowan, J, Notherm, B, Patt, M, Vue, L, Bianchetti, C.M. | | Deposit date: | 2015-09-04 | | Release date: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure of Tetragonal Lysozyme in complex with Iodine solved by UWO Students
To Be Published
|
|
