3BLJ
 
 | | Crystal structure of human poly(ADP-ribose) polymerase 15, catalytic fragment | | Descriptor: | CHLORIDE ION, GLYCEROL, Poly(ADP-ribose) polymerase 15, ... | | Authors: | Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-11 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
3C63
 
 | | Tetrameric Cytochrome cb562 (K34/H59/D62/H63/H73/A74/H77) Assembly Stabilized by Interprotein Zinc Coordination | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562, ... | | Authors: | Tezcan, F.A, Salgado, E.N, Lewis, R.A, Faraone-Mennella, J. | | Deposit date: | 2008-02-02 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Metal-mediated self-assembly of protein superstructures: influence of secondary interactions on protein oligomerization and aggregation.
J.Am.Chem.Soc., 130, 2008
|
|
3BU5
 
 | |
3BNO
 
 | |
2FI1
 
 | | The crystal structure of a hydrolase from Streptococcus pneumoniae TIGR4 | | Descriptor: | CALCIUM ION, hydrolase, haloacid dehalogenase-like family | | Authors: | Zhang, R, Zhou, M, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-27 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.4A crystal structure of a hydrolase from Streptococcus pneumoniae TIGR4
To be Published
|
|
3BS4
 
 | | Crystal structure of uncharacterized protein PH0321 from Pyrococcus horikoshii in complex with an unknown peptide | | Descriptor: | Uncharacterized protein PH0321, Unknown peptide | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of uncharacterized protein PH0321 from Pyrococcus horikoshii in complex with an unknown peptide.
To be Published
|
|
2FJY
 
 | |
3C19
 
 | | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein MK0293 | | Authors: | Patskovsky, Y, Romero, R, Bonanno, J.B, Malashkevich, V, Dickey, M, Chang, S, Koss, J, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19.
To be Published
|
|
3C1B
 
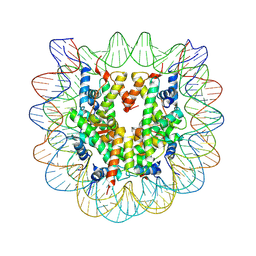 | | The effect of H3 K79 dimethylation and H4 K20 trimethylation on nucleosome and chromatin structure | | Descriptor: | Histone 2, H2bf, Histone H2A type 1, ... | | Authors: | Lu, X, Simon, M, Chodaparambil, J, Hansen, J, Shokat, K, Luger, K. | | Deposit date: | 2008-01-22 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2FLQ
 
 | |
3C3E
 
 | | Crystal structure of 2-phospho-(S)-lactate transferase from Methanosarcina mazei in complex with Fo and GDP. Northeast Structural Genomics Consortium target MaR46 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, 2-phospho-L-lactate transferase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Forouhar, F, Abashidze, M, Xu, H, Grochowski, L.L, Seetharaman, J, Hussain, M, Kuzin, A.P, Chen, Y, Zhou, W, Xiao, R, Acton, T.B, Montelione, G.T, Galinier, A, White, R.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular insights into the biosynthesis of the f420 coenzyme.
J.Biol.Chem., 283, 2008
|
|
2ESX
 
 | | The structure of the V3 region within gp120 of JR-FL HIV-1 strain (minimized average structure) | | Descriptor: | Envelope polyprotein GP160 | | Authors: | Rosen, O, Sharon, M, Samson, A.O, Quadt, S.R, Anglister, J. | | Deposit date: | 2005-10-27 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular switch for alternative conformations of the HIV-1 V3 region: Implications for phenotype conversion.
Proc.Natl.Acad.Sci.Usa, 13, 2006
|
|
2ESZ
 
 | | The structure of the V3 region within gp120 of JR-FL HIV-1 strain (ensemble) | | Descriptor: | Envelope polyprotein GP160 | | Authors: | Rosen, O, Sharon, M, Samson, A.O, Quadt, S.R, Anglister, J. | | Deposit date: | 2005-10-27 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular switch for alternative conformations of the HIV-1 V3 region: Implications for phenotype conversion.
Proc.Natl.Acad.Sci.Usa, 13, 2006
|
|
1LY9
 
 | | The impact of the physical and chemical environment on the molecular structure of Coprinus cinereus peroxidase | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase | | Authors: | Houborg, K, Harris, P, Petersen, J.F.W, Rowland, P, Poulsen, J.-C.N, Schneider, P, Vind, J, Larsen, S. | | Deposit date: | 2002-06-07 | | Release date: | 2002-06-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Impact of the physical and chemical environment on the molecular structure of Coprinus cinereus peroxidase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2EUG
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE AND ITS COMPLEXES WITH URACIL AND GLYCEROL: STRUCTURE AND GLYCOSYLASE MECHANISM REVISITED | | Descriptor: | PROTEIN (GLYCOSYLASE), URACIL | | Authors: | Xiao, G, Tordova, M, Jagadeesh, J, Drohat, A.C, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-10-13 | | Release date: | 1999-10-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
3C6F
 
 | | Crystal structure of protein Bsu07140 from Bacillus subtilis | | Descriptor: | GLYCEROL, YetF protein | | Authors: | Patskovsky, Y, Min, T, Zhang, A, Adams, J, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein Bsu07140 from Bacillus subtilis.
To be Published
|
|
3C4S
 
 | | Crystal structure of the Ssl0352 protein from Synechocystis sp. Northeast Structural Genomics Consortium target SgR42 | | Descriptor: | Ssl0352 protein | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Wang, D, Maglaqui, M, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Ssl0352 protein from Synechocystis sp.
To be Published
|
|
2EXE
 
 | | Crystal structure of the phosphorylated CLK3 | | Descriptor: | Dual specificity protein kinase CLK3 | | Authors: | Papagrigoriou, E, Rellos, P, Das, S, Bullock, A, Ball, L.J, Turnbull, A, Savitsky, P, Fedorov, O, Johansson, C, Ugochukwu, E, Sobott, F, von Delft, F, Edwards, A, Sundstrom, M, Weigelt, J, Arrowsmith, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-08 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the phosphorylated CLK3
TO BE PUBLISHED
|
|
3C97
 
 | | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae | | Descriptor: | Signal transduction histidine kinase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae.
To be Published
|
|
3BMQ
 
 | | Structure of Pteridine Reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor (NADP+) and inhibitor (Compound AX5) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-(benzylsulfanyl)pyrimidine-2,4-diamine, ACETATE ION, ... | | Authors: | Martini, V.P, Iulek, J, Hunter, W.N. | | Deposit date: | 2007-12-13 | | Release date: | 2008-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of pteridine reductase inhibitors targeting african sleeping sickness and the leishmaniases.
J.Med.Chem., 53, 2010
|
|
2FHQ
 
 | |
2FDH
 
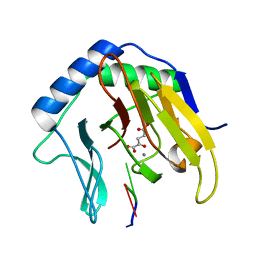 | | Crystal Structure of AlkB in complex with Mn(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(P*TP*(MA7)P*T)-3', Alkylated DNA repair protein alkB, ... | | Authors: | Yu, B, Benach, J, Edstrom, W.C, Gibney, B.R, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB.
Nature, 439, 2006
|
|
3BQW
 
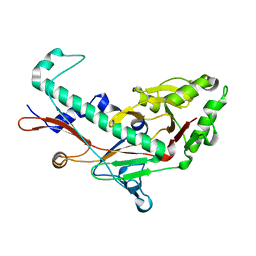 | |
1HH9
 
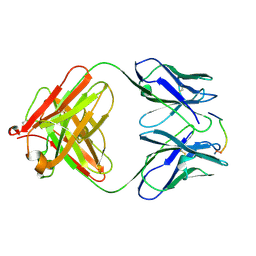 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE | | Descriptor: | IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN), IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN), PEP-2 | | Authors: | Hahn, M, Wessner, H, Schneider-Mergener, J, Hohne, W. | | Deposit date: | 2000-12-21 | | Release date: | 2001-01-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Evolutionary Transition Pathways for Changing Peptide Ligand Specificity and Structure
Embo J., 19, 2000
|
|
2ERS
 
 | |
