8FQK
 
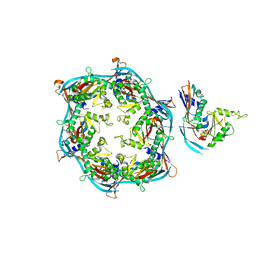 | | Asymmetric unit of HK97 phage prohead I | | Descriptor: | Scaffolding domain delta | | Authors: | Huet, A, Oh, B, Maurer, J, Duda, R.L, Conway, J.F. | | Deposit date: | 2023-01-06 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A symmetry mismatch unraveled: How phage HK97 scaffold flexibly accommodates a 12-fold pore at a 5-fold viral capsid vertex.
Sci Adv, 9, 2023
|
|
1BW5
 
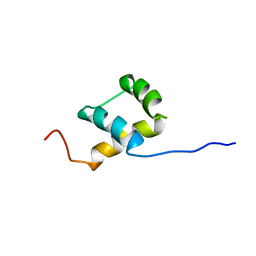 | | THE NMR SOLUTION STRUCTURE OF THE HOMEODOMAIN OF THE RAT INSULIN GENE ENHANCER PROTEIN ISL-1, 50 STRUCTURES | | Descriptor: | INSULIN GENE ENHANCER PROTEIN ISL-1 | | Authors: | Ippel, J.H, Larsson, G, Behravan, G, Zdunek, J, Lundqvist, M, Schleucher, J, Lycksell, P.-O, Wijmenga, S.S. | | Deposit date: | 1998-09-29 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the homeodomain of the rat insulin-gene enhancer protein isl-1. Comparison with other homeodomains.
J.Mol.Biol., 288, 1999
|
|
4XAP
 
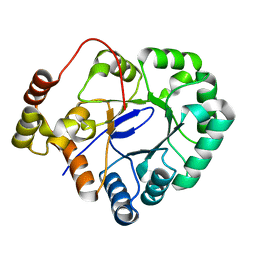 | | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 | | Descriptor: | Aldo-keto reductase | | Authors: | Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021
to be published
|
|
3R5Z
 
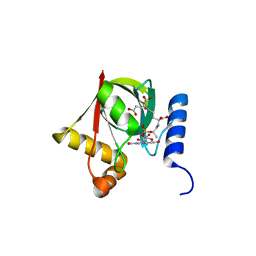 | | Structure of a Deazaflavin-dependent reductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
4XBS
 
 | | 2-deoxyribose-5-phosphate aldolase mutant - E78K | | Descriptor: | Deoxyribose-phosphate aldolase | | Authors: | Jiao, X.-C, Pan, J, Xu, G.-C, Kong, X.-D, Chen, Q, Zhang, Z.-J, Xu, J.-H. | | Deposit date: | 2014-12-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Efficient synthesis of a statin precursor in high space-time yield by a new aldehyde-tolerant aldolase identified from Lactobacillus brevis
Catalysis Science And Technology, 5, 2015
|
|
4XDD
 
 | | Apo [FeFe]-Hydrogenase CpI | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Esselborn, J, Hofmann, E, Kurisu, G, Happe, T. | | Deposit date: | 2014-12-19 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | A structural view of synthetic cofactor integration into [FeFe]-hydrogenases.
Chem Sci, 7, 2016
|
|
8FBG
 
 | | Crystal structure of NSD1 Mutant-Y1869C | | Descriptor: | CALCIUM ION, Histone-lysine N-methyltransferase, H3 lysine-36 specific, ... | | Authors: | Providokhina, K, Dong, A, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of NSD1
To Be Published
|
|
8FBH
 
 | | Crystal structure of NSD1 Mutant-T1927A | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Providokhina, K, Dong, A, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of NSD1
To Be Published
|
|
238L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-23 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
4D58
 
 | |
245L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
4J5Q
 
 | | TARG1 (C6orf130), Terminal ADP-ribose Glycohydrolase 1, apo structure | | Descriptor: | O-acetyl-ADP-ribose deacetylase 1 | | Authors: | Schellenberg, M.J, Appel, C.D, Krahn, J, Williams, R.S. | | Deposit date: | 2013-02-09 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease.
Embo J., 32, 2013
|
|
4XHS
 
 | | Crystal structure of human NLRP12 PYD domain and implication in homotypic interaction | | Descriptor: | FORMIC ACID, Maltose-binding periplasmic protein,NACHT, LRR and PYD domains-containing protein 12, ... | | Authors: | Jin, T, Huang, M, Jiang, J, Xiao, T. | | Deposit date: | 2015-01-06 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human NLRP12 PYD domain and implication in homotypic interaction
To Be Published
|
|
4XID
 
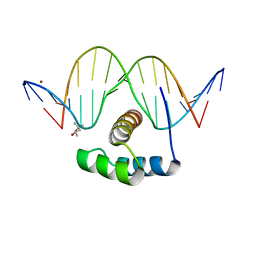 | | AntpHD with 15bp DNA duplex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J. | | Deposit date: | 2015-01-06 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Entropic Enhancement of Protein-DNA Affinity by Oxygen-to-Sulfur Substitution in DNA Phosphate.
Biophys.J., 109, 2015
|
|
1H9Y
 
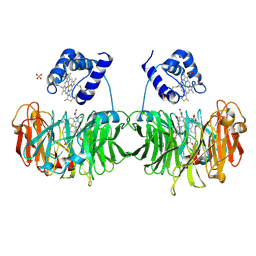 | |
8FHD
 
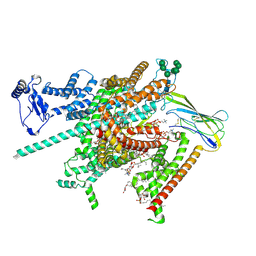 | | Cryo-EM structure of human voltage-gated sodium channel Nav1.6 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (5E,17R,20S)-23-amino-20-hydroxy-14,20-dioxo-15,19,21-trioxa-20lambda~5~-phosphatricos-5-en-17-yl hexadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of human voltage-gated sodium channel Na v 1.6.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5VEI
 
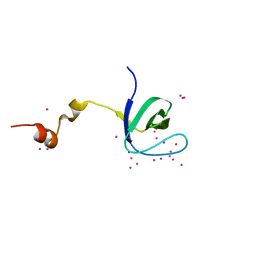 | | Crystal structure of the SH3 domain of human sorbin and SH3 domain-containing protein 2 | | Descriptor: | Sorbin and SH3 domain-containing protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Huang, H, Gu, J, Liu, K, Sidhu, S.S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal structure of the SH3 domain of human sorbin and SH3 domain-containing protein 2
To be Published
|
|
8FIS
 
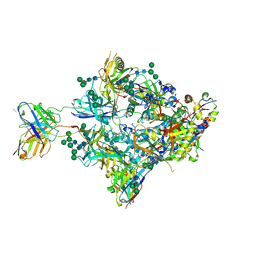 | | Structure of Bispecific CAP256V2LS-J3 Fab in complex with BG505 DS-SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-12-16 | | Release date: | 2023-02-01 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Bispecific antibody CAP256.J3LS targets V2-apex and CD4-binding sites with high breadth and potency.
Mabs, 15, 2023
|
|
1V1R
 
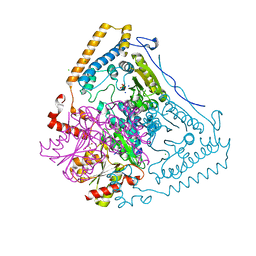 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, CHLORIDE ION, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-22 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain {Alpha}-Keto Acid Decarboxylase/Dehydrogenase
J.Biol.Chem., 279, 2004
|
|
4XO9
 
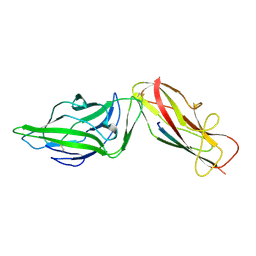 | | Crystal structure of a FimH*DsG complex from E.coli K12 in space group C2 | | Descriptor: | Minor component of type 1 fimbriae, Protein FimH | | Authors: | Jakob, R.P, Eras, J, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
240L
 
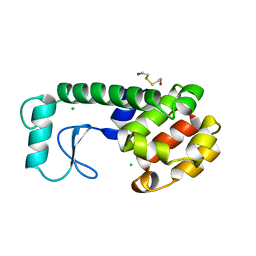 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
246L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
264D
 
 | | THREE-DIMENSIONAL CRYSTAL STRUCTURE OF THE A-TRACT DNA DODECAMER D(CGCAAATTTGCG) COMPLEXED WITH THE MINOR-GROOVE-BINDING DRUG HOECHST 33258 | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Vega, M.C, Garcia-Saez, I, Aymami, J, Eritja, R, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Coll, M. | | Deposit date: | 1994-09-22 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Three-dimensional crystal structure of the A-tract DNA dodecamer d(CGCAAATTTGCG) complexed with the minor-groove-binding drug Hoechst 33258.
Eur.J.Biochem., 222, 1994
|
|
8FJD
 
 | |
4X9O
 
 | | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae soaked with octanoyl-CoA: conformational changes without clearly bound substrate | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2 | | Authors: | Hou, J, Cooper, D.R, Shabalin, I.G, Grabowski, M, Shumilin, I, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and enzymatic studies of beta-ketoacyl-(acyl carrier protein) synthase III (FabH) from Vibrio cholerae
to be published
|
|
