1JF2
 
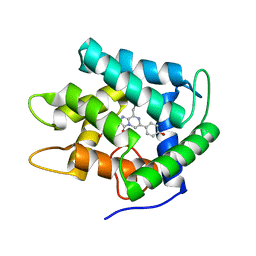 | | Crystal Structure of W92F obelin mutant from Obelia longissima at 1.72 Angstrom resolution | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, obelin | | Authors: | Liu, Z.-J, Vysotski, E.S, Deng, L, Markova, S.V, Lee, J, Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-06-19 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Violet bioluminescence and fast kinetics from W92F obelin: structure-based proposals for the bioluminescence triggering and the identification of the emitting species.
Biochemistry, 42, 2003
|
|
4ZB3
 
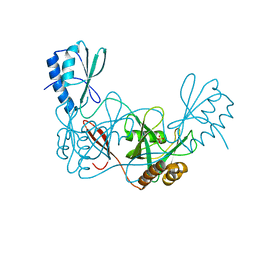 | | Crystal structure of the apo AtNUDT7 | | Descriptor: | Nudix hydrolase 7, SULFATE ION | | Authors: | Tang, Q, Liu, C, Zhong, C, Ding, J. | | Deposit date: | 2015-04-14 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana Nudix Hydrolase NUDT7 Reveal a Previously Unobserved Conformation.
Mol Plant, 8, 2015
|
|
4ZCL
 
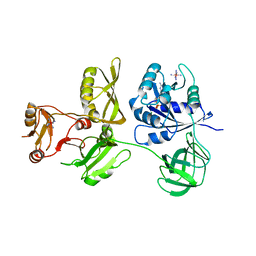 | | Crystal Structure of Escherichia coli GTPase BipA/TypA Complexed with GDP | | Descriptor: | COBALT HEXAMMINE(III), GTP-binding protein TypA/BipA, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fan, H.T, Hahm, J, Diggs, S, Blaha, G. | | Deposit date: | 2015-04-16 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural and Functional Analysis of BipA, a Regulator of Virulence in Enteropathogenic Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
4JQX
 
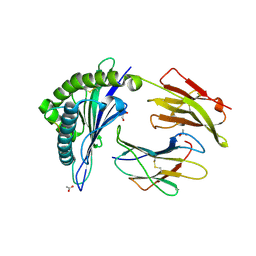 | | HLA-B*44:03 in complex with Epstein-Barr virus BZLF1-derived peptide (residues 169-180) | | Descriptor: | ACETATE ION, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Theodossis, A, Welland, A, Gras, S, Rossjohn, J. | | Deposit date: | 2013-03-20 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HLA Peptide Length Preferences Control CD8+ T Cell Responses.
J.Immunol., 191, 2013
|
|
4ZEJ
 
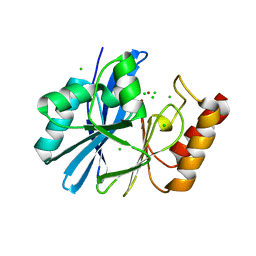 | |
4ZEV
 
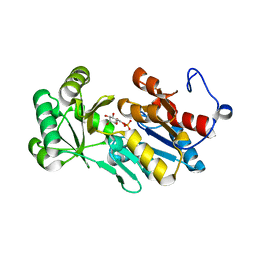 | | Crystal structure of PfHAD1 in complex with mannose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-mannopyranose, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2015-04-20 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cap-domain closure enables diverse substrate recognition by the C2-type haloacid dehalogenase-like sugar phosphatase Plasmodium falciparum HAD1.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
1H6T
 
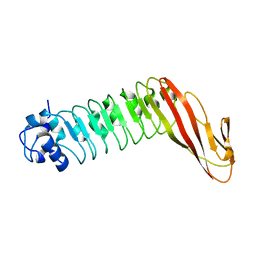 | | Internalin B: crystal structure of fused N-terminal domains. | | Descriptor: | INTERNALIN B | | Authors: | Schubert, W.-D, Gobel, G, Diepholz, M, Darji, A, Kloer, D, Hain, T, Chakraborty, T, Wehland, J, Domann, E, Heinz, D.W. | | Deposit date: | 2001-06-22 | | Release date: | 2001-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Internalins from the human pathogen Listeria monocytogenes combine three distinct folds into a contiguous internalin domain.
J.Mol.Biol., 312, 2001
|
|
4ZGO
 
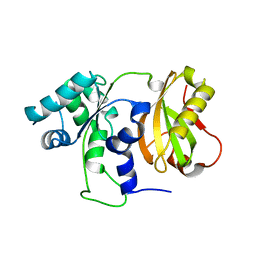 | | Structure of C-terminally truncated Cdc123 from Schizosaccharomyces pombe | | Descriptor: | Cell division cycle protein 123 | | Authors: | Panvert, M, Dubiez, E, Arnold, L, Perez, J, Seufert, W, Mechulam, Y, Schmitt, E. | | Deposit date: | 2015-04-23 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | Cdc123, a Cell Cycle Regulator Needed for eIF2 Assembly, Is an ATP-Grasp Protein with Unique Features.
Structure, 23, 2015
|
|
4ZH0
 
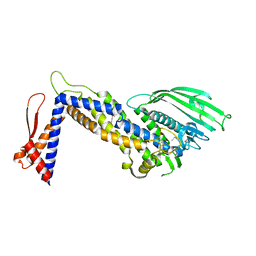 | | Structure of Helicobacter pylori adhesin BabA determined by SeMet SAD | | Descriptor: | Outer membrane protein-adhesin | | Authors: | Howard, T.D, Hage, N, Phillips, C, Brassington, C.A, Debreczeni, J, Overman, R, Gellert, P, Stolnik, S, Winkler, G.S, Falcone, F.H. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-19 | | Last modified: | 2015-12-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis of Lewis(b) antigen binding by the Helicobacter pylori adhesin BabA.
Sci Adv, 1, 2015
|
|
4WQF
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with elongation factor G and fusidic acid in the post-translocational state | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Lin, J, Gagnon, M.G, Steitz, T.A. | | Deposit date: | 2014-10-21 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Changes of Elongation Factor G on the Ribosome during tRNA Translocation.
Cell, 160, 2015
|
|
8ESK
 
 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with rocuronium, resting-like state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-10-14 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
8F6Y
 
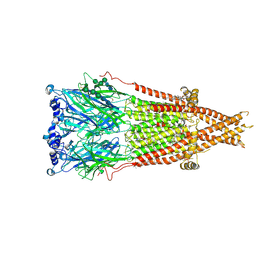 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with etomidate, desensitized-like state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-11-17 | | Release date: | 2023-06-07 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
8F6Z
 
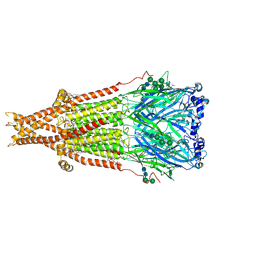 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with succinylcholine, desensitized-like state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2,2'-[(1,4-DIOXOBUTANE-1,4-DIYL)BIS(OXY)]BIS(N,N,N-TRIMETHYLETHANAMINIUM), 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-11-17 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
8F2S
 
 | | Cryo-EM structure of Torpedo nicotinic acetylcholine receptor in complex with rocuronium, pore-blocked state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goswami, U, Rahman, M.M, Teng, J, Hibbs, R.E. | | Deposit date: | 2022-11-08 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural interplay of anesthetics and paralytics on muscle nicotinic receptors.
Nat Commun, 14, 2023
|
|
4JXH
 
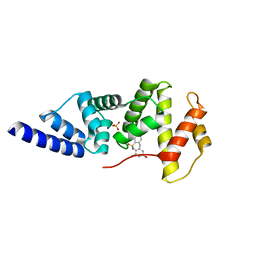 | |
4WFJ
 
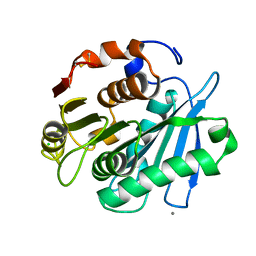 | | Crystal structure of PET-degrading cutinase Cut190 S226P mutant in Ca(2+)-bound state at 1.75 angstrom resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cutinase | | Authors: | Miyakawa, T, Mizushima, H, Ohtsuka, J, Oda, M, Kawai, F, Tanokura, M. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-24 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4WFK
 
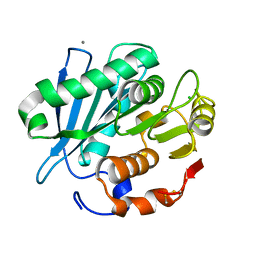 | | Crystal structure of PET-degrading cutinase Cut190 S226P mutant in Ca(2+)-bound state at 2.35 angstrom resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cutinase | | Authors: | Miyakawa, T, Mizushima, H, Ohtsuka, J, Oda, M, Kawai, F, Tanokura, M. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-24 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4WUO
 
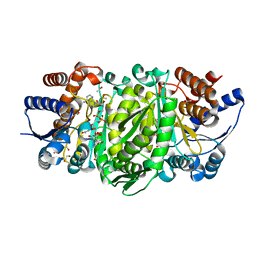 | | Structure of the E270A Mutant Isopropylmalate dehydrogenase from Thermus thermophilus in complex with IPM, Mn and NADH | | Descriptor: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase, ETHANOL, ... | | Authors: | Pallo, A, Graczer, E, Olah, J, Szimler, T, Konarev, P.V, Svergun, D.I, Merli, A, Zavodszky, P, Vas, M, Weiss, M.S. | | Deposit date: | 2014-11-03 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Glutamate 270 plays an essential role in K(+)-activation and domain closure of Thermus thermophilus isopropylmalate dehydrogenase.
Febs Lett., 589, 2015
|
|
4WUZ
 
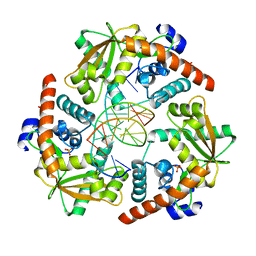 | | Crystal structure of lambda exonuclease in complex with DNA and Ca2+ | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*T*TP*CP*GP*GP*TP*AP*CP*AP*GP*TP*AP*G)-3'), DNA (5'-D(P*AP*GP*CP*TP*AP*CP*TP*GP*TP*AP*CP*CP*GP*A)-3'), ... | | Authors: | Zhang, J, Bell, C.E. | | Deposit date: | 2014-11-04 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of lambda Exonuclease in Complex with DNA and Ca(2+).
Biochemistry, 53, 2014
|
|
1YI3
 
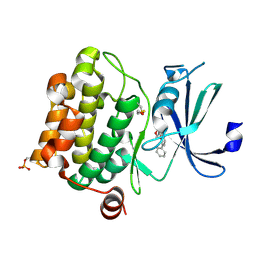 | | Crystal Structure of Pim-1 bound to LY294002 | | Descriptor: | 2-MORPHOLIN-4-YL-7-PHENYL-4H-CHROMEN-4-ONE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Jacobs, M.D, Black, J, Futer, O, Swenson, L, Hare, B, Fleming, M, Saxena, K. | | Deposit date: | 2005-01-11 | | Release date: | 2005-01-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pim-1 ligand-bound structures reveal the mechanism of serine/threonine kinase inhibition by LY294002.
J.Biol.Chem., 280, 2005
|
|
4F7C
 
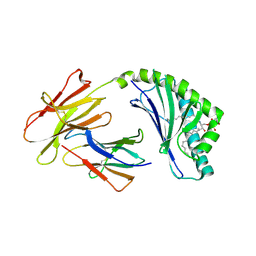 | | Crystal structure of bovine CD1d with bound C12-di-sulfatide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, CD1D antigen, ... | | Authors: | Wang, J, Zajonc, D.M. | | Deposit date: | 2012-05-15 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | Crystal Structures of Bovine CD1d Reveal Altered αGalCer Presentation and a Restricted A' Pocket Unable to Bind Long-Chain Glycolipids.
Plos One, 7, 2012
|
|
4WK6
 
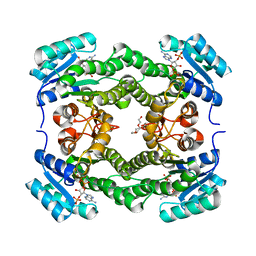 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) (G141A) from Vibrio cholerae in complex with NADPH | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL | | Authors: | Hou, J, Zheng, H, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
1RSC
 
 | | STRUCTURE OF AN EFFECTOR INDUCED INACTIVATED STATE OF RIBULOSE BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE: THE BINARY COMPLEX BETWEEN ENZYME AND XYLULOSE BISPHOSPHATE | | Descriptor: | RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN), XYLULOSE-1,5-BISPHOSPHATE | | Authors: | Newman, J, Gutteridge, S. | | Deposit date: | 1994-03-29 | | Release date: | 1995-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of an effector-induced inactivated state of ribulose 1,5-bisphosphate carboxylase/oxygenase: the binary complex between enzyme and xylulose 1,5-bisphosphate.
Structure, 2, 1994
|
|
4J87
 
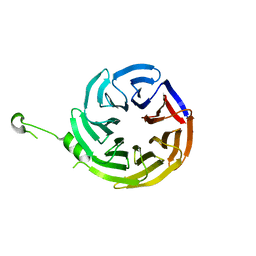 | | Crystal structure of alpha-COP | | Descriptor: | coatomer subunit alpha | | Authors: | Ma, W, Goldberg, J. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Rules for the recognition of dilysine retrieval motifs by coatomer.
Embo J., 32, 2013
|
|
1RTJ
 
 | | MECHANISM OF INHIBITION OF HIV-1 REVERSE TRANSCRIPTASE BY NON-NUCLEOSIDE INHIBITORS | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Ross, C, Jones, Y, Stammers, D, Stuart, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
