6PS4
 
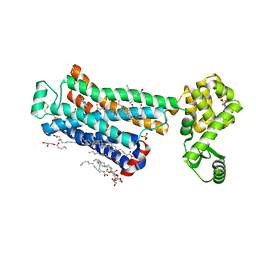 | | XFEL beta2 AR structure by ligand exchange from Timolol to ICI-118551. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3S)-1-[(7-methyl-2,3-dihydro-1H-inden-4-yl)oxy]-3-[(1-methylethyl)amino]butan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
6PS5
 
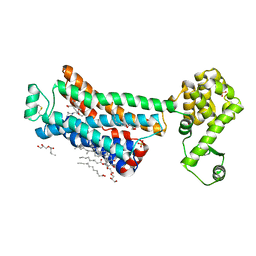 | | XFEL beta2 AR structure by ligand exchange from Timolol to Propranolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(ISOPROPYLAMINO)-3-(1-NAPHTHYLOXY)-2-PROPANOL, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
8XI6
 
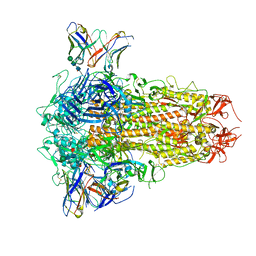 | | SARS-CoV-2 Omicron BQ.1.1 Variant Spike Protein Complexed with MO11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ishimaru, H, Nishimura, M, Shigematsu, H, Marini, M.I, Hasegawa, N, Takamiya, R, Iwata, S, Mori, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Epitopes of an antibody that neutralizes a wide range of SARS-CoV-2 variants in a conserved subdomain 1 of the spike protein.
J.Virol., 98, 2024
|
|
1KQG
 
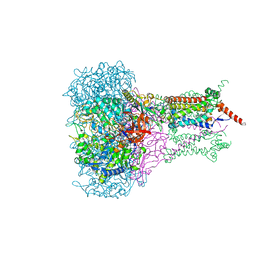 | | FORMATE DEHYDROGENASE N FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, CARDIOLIPIN, ... | | Authors: | Jormakka, M, Tornroth, S, Byrne, B, Iwata, S. | | Deposit date: | 2002-01-05 | | Release date: | 2002-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of proton motive force generation: structure of formate dehydrogenase-N.
Science, 295, 2002
|
|
4AU5
 
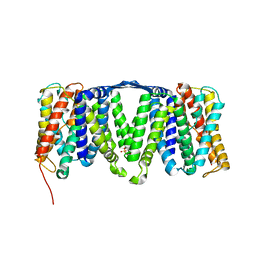 | | Structure of the NhaA dimer, crystallised at low pH | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, NA(+)/H(+) ANTIPORTER NHAA, SULFATE ION | | Authors: | Drew, D, Lee, C, Iwata, S, Cameron, A.D. | | Deposit date: | 2012-05-14 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.696 Å) | | Cite: | Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights.
J. Gen. Physiol., 144, 2014
|
|
1KQF
 
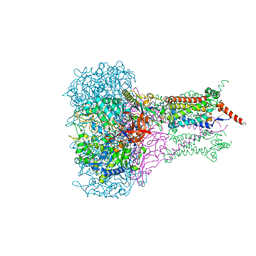 | | FORMATE DEHYDROGENASE N FROM E. COLI | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CARDIOLIPIN, FORMATE DEHYDROGENASE, ... | | Authors: | Jormakka, M, Tornroth, S, Byrne, B, Iwata, S. | | Deposit date: | 2002-01-05 | | Release date: | 2002-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of proton motive force generation: structure of formate dehydrogenase-N.
Science, 295, 2002
|
|
4CAD
 
 | | Mechanism of farnesylated CAAX protein processing by the integral membrane protease Rce1 | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kulkarni, K, Manolaridis, I, Dodd, R.B, Cronin, N, Ogasawara, S, Iwata, S, Barford, D. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Farnesylated Caax Protein Processing by the Intramembrane Protease Rce1
Nature, 504, 2013
|
|
6PS6
 
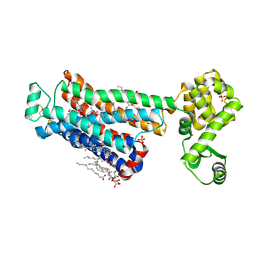 | | XFEL beta2 AR structure by ligand exchange from Timolol to Timolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(tert-butylamino)-3-[(4-morpholin-4-yl-1,2,5-thiadiazol-3-yl)oxy]propan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
2V8N
 
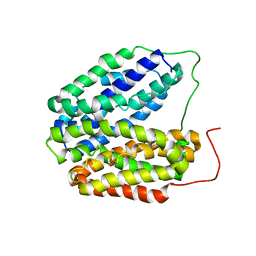 | | Wild-type Structure of Lactose Permease | | Descriptor: | LACTOSE PERMEASE | | Authors: | Guan, L, Mirza, O, Verner, G, Iwata, S, Kaback, H.R. | | Deposit date: | 2007-08-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Determination of Wild-Type Lactose Permease.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2VOC
 
 | | THIOREDOXIN A ACTIVE SITE MUTANTS FORM MIXED DISULFIDE DIMERS THAT RESEMBLE ENZYME-SUBSTRATE REACTION INTERMEDIATE | | Descriptor: | DI(HYDROXYETHYL)ETHER, THIOREDOXIN | | Authors: | Kouwen, T.R.H.M, Andrell, J, Schrijver, R, Dubois, J.Y.F, Maher, M.J, Iwata, S, Carpenter, E.P, van Dijl, J.M. | | Deposit date: | 2008-02-13 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Thioredoxin A active-site mutants form mixed disulfide dimers that resemble enzyme-substrate reaction intermediates.
J. Mol. Biol., 379, 2008
|
|
2D04
 
 | | Crystal structure of neoculin, a sweet protein with taste-modifying activity. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Curculin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shimizu-Ibuka, A, Morita, Y, Terada, T, Asakura, T, Nakajima, K, Iwata, S, Misaka, T, Sorimachi, H, Arai, S, Abe, K. | | Deposit date: | 2005-07-25 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structure of neoculin: insights into its sweetness and taste-modifying activity
J.Mol.Biol., 359, 2006
|
|
4D1C
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH bromovinylhydantoin bound. | | Descriptor: | (5Z)-5-[(3-bromophenyl)methylidene]imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
8WZV
 
 | | SFX structure of an Mn-carbonyl complex immobilized in hen egg white lysozyme microcrystals, 1 microsecond after photoexcitation with 40 microJoules laser intensity at RT. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Maity, B, Shoji, M, Luo, F, Nakane, T, Abe, S, Owada, S, Kang, J, Tono, K, Tanaka, R, Thuc, T.T, Kojima, M, Hishikawa, Y, Tanaka, J, Tian, J, Noya, H, Nakasuji, Y, Asanuma, A, Yao, X, Iwata, S, Shigeta, Y, Nango, E, Ueno, T. | | Deposit date: | 2023-11-02 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Real-time observation of a metal complex-driven reaction intermediate using a porous protein crystal and serial femtosecond crystallography.
Nat Commun, 15, 2024
|
|
8WZT
 
 | | SFX structure of an Mn-carbonyl complex immobilized in hen egg white lysozyme microcrystals, 1 microsecond after photoexcitation at RT. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Maity, B, Shoji, M, Luo, F, Nakane, T, Abe, S, Owada, S, Kang, J, Tono, K, Tanaka, R, Thuc, T.T, Kojima, M, Hishikawa, Y, Tanaka, J, Tian, J, Noya, H, Nakasuji, Y, Asanuma, A, Yao, X, Iwata, S, Shigeta, Y, Nango, E, Ueno, T. | | Deposit date: | 2023-11-02 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Real-time observation of a metal complex-driven reaction intermediate using a porous protein crystal and serial femtosecond crystallography.
Nat Commun, 15, 2024
|
|
8WZF
 
 | | Dark state SFX structure of hen egg white lysozyme microcrystals immobilized with a Mn carbonyl complex at RT. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Maity, B, Shoji, M, Luo, F, Nakane, T, Abe, S, Owada, S, Kang, J, Tono, K, Tanaka, R, Thuc, T.T, Kojima, M, Hishikawa, Y, Tanaka, J, Tian, J, Noya, H, Nakasuji, Y, Asanuma, A, Yao, X, Iwata, S, Shigeta, Y, Nango, E, Ueno, T. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Real-time observation of a metal complex-driven reaction intermediate using a porous protein crystal and serial femtosecond crystallography.
Nat Commun, 15, 2024
|
|
8WZG
 
 | | SFX structure of an Mn-carbonyl complex immobilized in hen egg white lysozyme microcrystals, 10 ns after photoexcitation at RT. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Maity, B, Shoji, M, Luo, F, Nakane, T, Abe, S, Owada, S, Kang, J, Tono, K, Tanaka, R, Thuc, T.T, Kojima, M, Hishikawa, Y, Tanaka, J, Tian, J, Noya, H, Nakasuji, Y, Asanuma, A, Yao, X, Iwata, S, Shigeta, Y, Nango, E, Ueno, T. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Real-time observation of a metal complex-driven reaction intermediate using a porous protein crystal and serial femtosecond crystallography.
Nat Commun, 15, 2024
|
|
4D1D
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER with the inhibitor 5-(2-naphthylmethyl)-L-hydantoin. | | Descriptor: | 5-(2-NAPHTHYLMETHYL)-D-HYDANTOIN, 5-(2-NAPHTHYLMETHYL)-L-HYDANTOIN, HYDANTOIN TRANSPORT PROTEIN, ... | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1B
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH BENZYL-HYDANTOIN | | Descriptor: | (5S)-5-benzylimidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Brueckner, F, Geng, T, Weyand, S, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1A
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH INDOLYLMETHYL-HYDANTOIN | | Descriptor: | (5S)-5-(1H-indol-3-ylmethyl)imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
8WZR
 
 | | SFX structure of an Mn-carbonyl complex immobilized in hen egg white lysozyme microcrystals, 100 ns after photoexcitation at RT. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Maity, B, Shoji, M, Luo, F, Nakane, T, Abe, S, Owada, S, Kang, J, Tono, K, Tanaka, R, Thuc, T.T, Kojima, M, Hishikawa, Y, Tanaka, J, Tian, J, Noya, H, Nakasuji, Y, Asanuma, A, Yao, X, Iwata, S, Shigeta, Y, Nango, E, Ueno, T. | | Deposit date: | 2023-11-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Real-time observation of a metal complex-driven reaction intermediate using a porous protein crystal and serial femtosecond crystallography.
Nat Commun, 15, 2024
|
|
7XRR
 
 | | Crystal structure of the human OX2R bound to the insomnia drug lemborexant. | | Descriptor: | (1~{R},2~{S})-2-[(2,4-dimethylpyrimidin-5-yl)oxymethyl]-~{N}-(5-fluoranylpyridin-2-yl)-2-(3-fluorophenyl)cyclopropane-1-carboxamide, Orexin receptor type 2 | | Authors: | Asada, H, Im, D, Iwata, S. | | Deposit date: | 2022-05-11 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Molecular basis for anti-insomnia drug design from structure of lemborexant-bound orexin 2 receptor.
Structure, 30, 2022
|
|
8WG4
 
 | | mouse TMEM63b in DDM-CHS micelle with YN9303-24 Fab | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL HEMISUCCINATE, CSC1-like protein 2,Green fluorescent protein | | Authors: | Miyata, Y, Takahashi, K, Lee, Y, Sultan, C.S, Kuribayashi, R, Takahashi, M, Hata, K, Bamba, T, Izumi, Y, Liu, K, Uemura, T, Nomura, N, Iwata, S, Nagata, S, Nishizawa, T, Segawa, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanosensitive channel TMEM63B functions as a plasma membrane lipid scramblase
To Be Published
|
|
8WG3
 
 | | mouse TMEM63b in LMNG-CHS micelle | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL HEMISUCCINATE, CSC1-like protein 2,Green fluorescent protein | | Authors: | Miyata, Y, Takahashi, K, Lee, Y, Sultan, C.S, Kuribayashi, R, Takahashi, M, Hata, K, Bamba, T, Izumi, Y, Liu, K, Uemura, T, Nomura, N, Iwata, S, Nagata, S, Nishizawa, T, Segawa, K. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanosensitive channel TMEM63B functions as a plasma membrane lipid scramblase
To Be Published
|
|
1Y43
 
 | | crystal structure of aspergilloglutamic peptidase from Aspergillus niger | | Descriptor: | Aspergillopepsin II heavy chain, Aspergillopepsin II light chain, SULFATE ION | | Authors: | Sasaki, H, Nakagawa, A, Iwata, S, Muramatsu, T, Suganuma, M, Sawano, Y, Kojima, M, Kubota, K, Takahashi, K. | | Deposit date: | 2004-11-30 | | Release date: | 2005-12-13 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The three-dimensional structure of aspergilloglutamic peptidase from Aspergillus niger
Proc.Jpn.Acad.,Ser.B, 80, 2004
|
|
4YSD
 
 | | Room temperature structure of copper nitrite reductase from Geobacillus thermodenitrificans | | Descriptor: | ACETIC ACID, COPPER (II) ION, Nitrite reductase | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
