4TQV
 
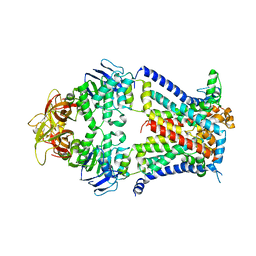 | | Crystal structure of a bacterial ABC transporter involved in the import of the acidic polysaccharide alginate | | Descriptor: | AlgM1, AlgM2, AlgS | | Authors: | Maruyama, Y, Itoh, T, Kaneko, A, Nishitani, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2014-06-12 | | Release date: | 2015-07-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.504 Å) | | Cite: | Structure of a Bacterial ABC Transporter Involved in the Import of an Acidic Polysaccharide Alginate
Structure, 23, 2015
|
|
7WOX
 
 | | PPARgamma antagonist (MMT-160)- PPARgamma LBD complex | | Descriptor: | N-[[5-(3-phenylprop-2-ynoylamino)-2-propoxy-phenyl]methyl]-4-pyrimidin-2-yl-benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Yoshizawa, M, Aoyama, T, Itoh, T, Miyachi, H. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Arylalkynyl amide-type peroxisome proliferator-activated receptor gamma (PPAR gamma )-selective antagonists covalently bind to the PPAR gamma ligand binding domain with a unique binding mode.
Bioorg.Med.Chem.Lett., 64, 2022
|
|
3K3T
 
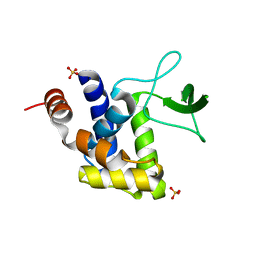 | | E185A mutant of peptidoglycan hydrolase from Sphingomonas sp. A1 | | Descriptor: | Peptidoglycan hydrolase FlgJ, SULFATE ION | | Authors: | Maruyama, Y, Ochiai, A, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-10-04 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mutational studies of the peptidoglycan hydrolase FlgJ of Sphingomonas sp. strain A1
J.Basic Microbiol., 50, 2010
|
|
5WR0
 
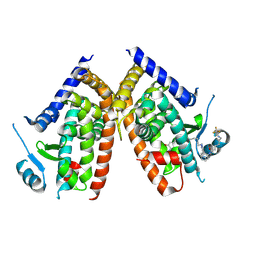 | | Huisgen cycloaddition for PPARg-LBD labeling by soaking method | | Descriptor: | (E)-N-[(3E)-2-oxo-16-(8-{6-[(trifluoroacetyl)amino]hexanoyl}-8,9-dihydro-1H-dibenzo[b,f][1,2,3]triazolo[4,5-d]azocin-1-yl)hexadec-3-en-1-ylidene]glycine, Peroxisome proliferator-activated receptor gamma | | Authors: | Kojima, H, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-11-29 | | Release date: | 2017-11-22 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | On-site reaction for PPAR gamma modification using a specific bifunctional ligand
Bioorg. Med. Chem., 25, 2017
|
|
5WR1
 
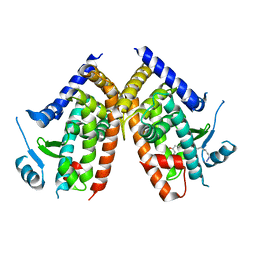 | | Covalent bond formation of bifunctional ligand with hPPARg-LBD | | Descriptor: | 2-[E-(E-16-azido-2-oxidanylidene-hexadec-3-enylidene)amino]ethanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Kojima, H, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-11-29 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | On-site reaction for PPAR gamma modification using a specific bifunctional ligand
Bioorg. Med. Chem., 25, 2017
|
|
3E9I
 
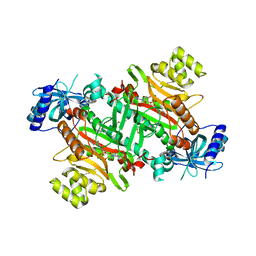 | | Lysyl-tRNA synthetase from Bacillus stearothermophilus complexed with L-Lysine hydroxamate-AMP | | Descriptor: | 5'-O-{(R)-hydroxy[(L-lysylamino)oxy]phosphoryl}adenosine, Lysyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sakurama, H, Takita, T, Mikami, B, Itoh, T, Yasukawa, K, Inouye, K. | | Deposit date: | 2008-08-22 | | Release date: | 2009-07-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two crystal structures of lysyl-tRNA synthetase from Bacillus stearothermophilus in complex with lysyladenylate-like compounds: insights into the irreversible formation of the enzyme-bound adenylate of L-lysine hydroxamate
J.Biochem., 145, 2009
|
|
3E9H
 
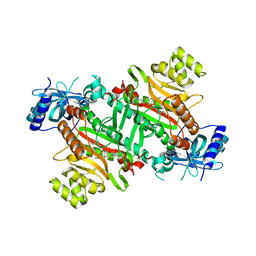 | | Lysyl-tRNA synthetase from Bacillus stearothermophilus complexed with L-Lysylsulfamoyl adenosine | | Descriptor: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Lysyl-tRNA synthetase, MAGNESIUM ION | | Authors: | Sakurama, H, Takita, T, Mikami, B, Itoh, T, Yasukawa, K, Inouye, K. | | Deposit date: | 2008-08-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two crystal structures of lysyl-tRNA synthetase from Bacillus stearothermophilus in complex with lysyladenylate-like compounds: insights into the irreversible formation of the enzyme-bound adenylate of L-lysine hydroxamate
J.Biochem., 145, 2009
|
|
5H1E
 
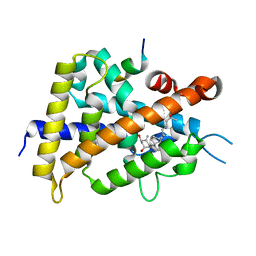 | | Interaction between vitamin D receptor and coactivator peptide SRC2-3 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Nuclear receptor coactivator 2 peptide, Vitamin D3 receptor | | Authors: | Egawa, D, Itoh, T, Kato, A, Kataoka, S, Anami, Y, Yamamoto, K. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | SRC2-3 binds to vitamin D receptor with high sensitivity and strong affinity
Bioorg. Med. Chem., 25, 2017
|
|
8HHP
 
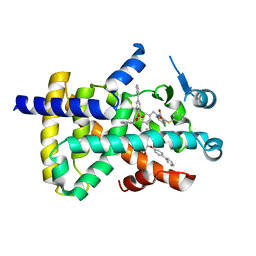 | |
8HHQ
 
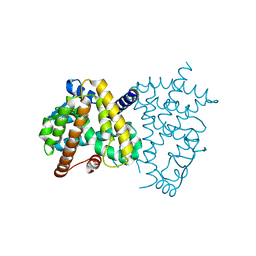 | |
2MWI
 
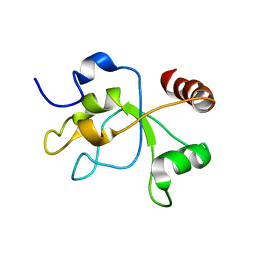 | | The structure of the carboxy-terminal domain of DNTTIP1 | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1 | | Authors: | Schwabe, J.W.R, Muskett, F.W, Itoh, T. | | Deposit date: | 2014-11-11 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of a cell cycle associated HDAC1/2 complex reveals the structural basis for complex assembly and nucleosome targeting.
Nucleic Acids Res., 43, 2015
|
|
5AYJ
 
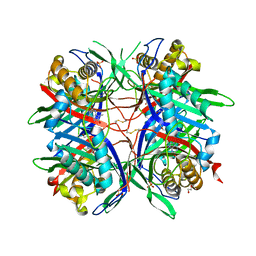 | | Hyperthermostable mutant of Bacillus sp. TB-90 Urate Oxidase - R298C | | Descriptor: | 9-METHYL URIC ACID, HEXAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Hibi, T, Kume, A, Kawamura, A, Itoh, T, Nishiya, Y. | | Deposit date: | 2015-08-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Hyperstabilization of Tetrameric Bacillus sp. TB-90 Urate Oxidase by Introducing Disulfide Bonds through Structural Plasticity
Biochemistry, 55, 2016
|
|
7CMQ
 
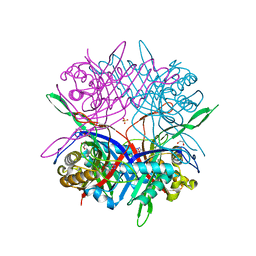 | |
7CUF
 
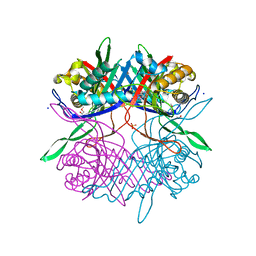 | |
7CMN
 
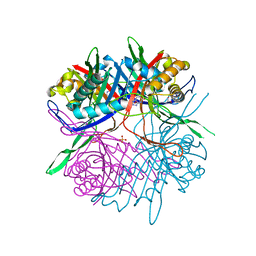 | |
7CUG
 
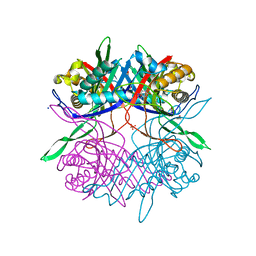 | |
5YJ2
 
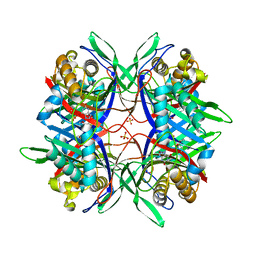 | |
5YJA
 
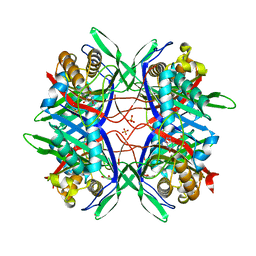 | |
5Z2B
 
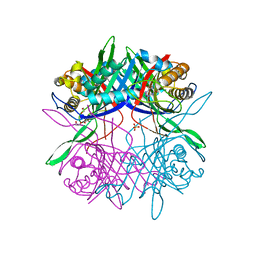 | |
5Z27
 
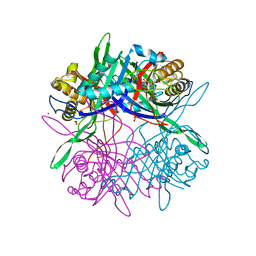 | |
7D9W
 
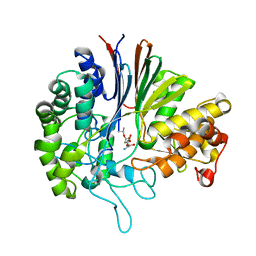 | | Gamma-glutamyltranspeptidase from Pseudomonas nitroreducens complexed with L-DON | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, GLYCINE, Gamma-glutamyltransferase 1 Threonine peptidase. MEROPS family T03 | | Authors: | Hibi, T, Sano, C, Putthapong, P, Hayashi, J, Itoh, T, Wakayama, M. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutagenesis and structure-based analysis of the role of Tryptophan525 of gamma-glutamyltranspeptidase from Pseudomonas nitroreducens.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7D9X
 
 | | Highly active mutant W525D of Gamma-glutamyltranspeptidase from Pseudomonas nitroreducens | | Descriptor: | GAMMA-BUTYROLACTONE, GLYCEROL, GLYCINE, ... | | Authors: | Hibi, T, Sano, C, Itoh, T, Wakayama, M. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mutagenesis and structure-based analysis of the role of Tryptophan525 of gamma-glutamyltranspeptidase from Pseudomonas nitroreducens.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7D9E
 
 | | Gamma-glutamyltranspeptidase from Pseudomonas nitroreducens complexed with L-DON | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, GLYCEROL, Gamma-glutamyltransferase 1 Threonine peptidase. MEROPS family T03 | | Authors: | Hibi, T, Sano, C, Itoh, T, Wakayama, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenesis and structure-based analysis of the role of Tryptophan525 of gamma-glutamyltranspeptidase from Pseudomonas nitroreducens.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
5AZT
 
 | | Ternary complex of hPPARalpha ligand binding domain, 17-oxoDHA and a SRC1 peptide | | Descriptor: | (4~{Z},7~{Z},10~{Z},13~{Z},19~{Z})-17-oxidanylidenedocosa-4,7,10,13,19-pentaenoic acid, 15-meric peptide from Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor alpha | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2015-10-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | 17-OxoDHA Is a PPAR alpha/gamma Dual Covalent Modifier and Agonist
Acs Chem.Biol., 2016
|
|
7CUC
 
 | |
