3K3T
 
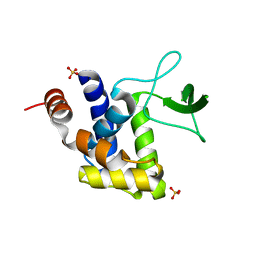 | | E185A mutant of peptidoglycan hydrolase from Sphingomonas sp. A1 | | Descriptor: | Peptidoglycan hydrolase FlgJ, SULFATE ION | | Authors: | Maruyama, Y, Ochiai, A, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-10-04 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mutational studies of the peptidoglycan hydrolase FlgJ of Sphingomonas sp. strain A1
J.Basic Microbiol., 50, 2010
|
|
5XPL
 
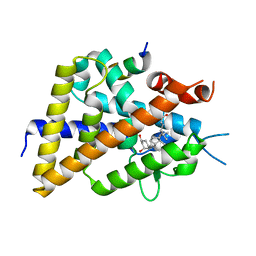 | | Crystal structure of VDR-LBD complexed with 22S-butyl-25-hydroxyphenyl-2-methylidene-19,26,27-trinor-25-oxo-1-hydroxyvitamin D3 | | Descriptor: | (4~{S})-4-[(1~{R})-1-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl )cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]ethyl]-1-(4-hydroxyphenyl)octan-1-one, Nuclear receptor coactivator 2, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
5XPM
 
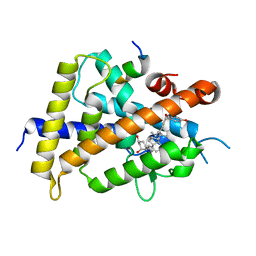 | | Crystal structure of VDR-LBD complexed with 22S-Butyl-25RS-(hydroxyphenyl)-25-methoxy-2-methylidene-19,26,27-trinor-1-hydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},3~{S})-3-[(3~{S})-3-(4-hydroxyphenyl)-3-methoxy-propyl]heptan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
5XPN
 
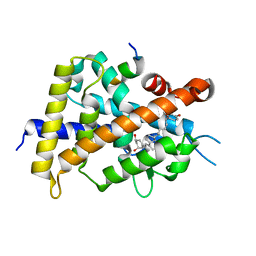 | | Crystal structure of VDR-LBD complexed with 25RS-(hydroxyphenyl)-25-methoxy-2-methylidene-19,26,27-trinor-1-hydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{R})-6-(4-hydroxyphenyl)-6-methoxy-hexan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{S})-6-(4-hydroxyphenyl)-6-methoxy-hexan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
5XPP
 
 | | Crystal structure of VDR-LBD complexed with 25RS-(Hydroxyphenyl)-2-methylidene-19,26,27-trinor-1,25-dihydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{R})-6-(4-hydroxyphenyl)-6-oxidanyl-hexan-2-yl]-7~{ a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
5XPO
 
 | | Crystal structure of VDR-LBD complexed with 25-(hydroxyphenyl)-2-methylidene-19,26,27-trinor-25-oxo-1-hydroxyvitamin D3 | | Descriptor: | (5~{R})-5-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[2-[(3~{R},5~{R})-4-methylidene-3,5-bis(oxidanyl)cyclohexyl idene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]-1-(4-hydroxyphenyl)hexan-1-one, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
6JEY
 
 | | Covalent bond formation between ynone moiety of synthetic fatty acid and hPPARg-LBD | | Descriptor: | (9Z,12Z,15Z,18Z,21Z)-5-oxidanylidenetetracosa-9,12,15,18,21-pentaen-6-ynoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Kojima, H, Yamamoto, K, Itoh, T. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cyclization Reaction-Based Turn-on Probe for Covalent Labeling of Target Proteins.
Cell Chem Biol, 27, 2020
|
|
6JEZ
 
 | | Covalent labeling of rVDR-LBD by turn-on fluorescent probe mediated by conjugate addition and cyclization | | Descriptor: | (1R,3R)-5-(2-((1R,3aS,7aR,E)-1-((R)-6-hydroxy-6-methylheptan-2-yl)-7a-methyloctahydro-4H-inden-4-ylidene)ethylidene)-2- methylenecyclohexane-1,3-diol, 7-(diethylamino)chromen-2-one, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Kojima, H, Yamamoto, K, Itoh, T. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cyclization Reaction-Based Turn-on Probe for Covalent Labeling of Target Proteins.
Cell Chem Biol, 27, 2020
|
|
6JF0
 
 | |
6IQ5
 
 | | Crystal Structure of CYP1B1 and Inhibitor Having Azide Group | | Descriptor: | 2-(cis-4-azidocyclohexyl)-4H-naphtho[1,2-b]pyran-4-one, Cytochrome P450 1B1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kubo, M, Yamamoto, K, Itoh, T. | | Deposit date: | 2018-11-06 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Design and synthesis of selective CYP1B1 inhibitor via dearomatization of alpha-naphthoflavone.
Bioorg. Med. Chem., 27, 2019
|
|
7WOX
 
 | | PPARgamma antagonist (MMT-160)- PPARgamma LBD complex | | Descriptor: | N-[[5-(3-phenylprop-2-ynoylamino)-2-propoxy-phenyl]methyl]-4-pyrimidin-2-yl-benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Yoshizawa, M, Aoyama, T, Itoh, T, Miyachi, H. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Arylalkynyl amide-type peroxisome proliferator-activated receptor gamma (PPAR gamma )-selective antagonists covalently bind to the PPAR gamma ligand binding domain with a unique binding mode.
Bioorg.Med.Chem.Lett., 64, 2022
|
|
7CMQ
 
 | |
7CUF
 
 | |
7D9W
 
 | | Gamma-glutamyltranspeptidase from Pseudomonas nitroreducens complexed with L-DON | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, GLYCINE, Gamma-glutamyltransferase 1 Threonine peptidase. MEROPS family T03 | | Authors: | Hibi, T, Sano, C, Putthapong, P, Hayashi, J, Itoh, T, Wakayama, M. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutagenesis and structure-based analysis of the role of Tryptophan525 of gamma-glutamyltranspeptidase from Pseudomonas nitroreducens.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7D9X
 
 | | Highly active mutant W525D of Gamma-glutamyltranspeptidase from Pseudomonas nitroreducens | | Descriptor: | GAMMA-BUTYROLACTONE, GLYCEROL, GLYCINE, ... | | Authors: | Hibi, T, Sano, C, Itoh, T, Wakayama, M. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mutagenesis and structure-based analysis of the role of Tryptophan525 of gamma-glutamyltranspeptidase from Pseudomonas nitroreducens.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7D9E
 
 | | Gamma-glutamyltranspeptidase from Pseudomonas nitroreducens complexed with L-DON | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, GLYCEROL, Gamma-glutamyltransferase 1 Threonine peptidase. MEROPS family T03 | | Authors: | Hibi, T, Sano, C, Itoh, T, Wakayama, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenesis and structure-based analysis of the role of Tryptophan525 of gamma-glutamyltranspeptidase from Pseudomonas nitroreducens.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7E2O
 
 | | X-ray Crystal structure of PPARgamma R288H mutant. | | Descriptor: | Peroxisome proliferator-activated receptor gamma | | Authors: | Egawa, D, Itoh, T. | | Deposit date: | 2021-02-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights into the Loss-of-Function R288H Mutant of Human PPAR gamma.
Biol.Pharm.Bull., 44, 2021
|
|
7CUC
 
 | |
7CMN
 
 | |
3VW4
 
 | | Crystal structure of the DNA-binding domain of ColE2-P9 Rep in complex with the replication origin | | Descriptor: | DNA (5'-D(P*AP*AP*TP*GP*AP*GP*AP*CP*CP*AP*GP*AP*TP*AP*AP*GP*CP*CP*TP*TP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*AP*AP*GP*GP*CP*TP*TP*AP*TP*CP*TP*GP*GP*TP*CP*TP*CP*AP*TP*T)-3'), Rep, ... | | Authors: | Itou, H, Yagura, M, Itoh, T, Shirakihara, Y. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Replication Origin Unwinding by An Initiator-Primase of Plasmid ColE2-P9: Duplex DNA Unwinding by A Single Protein
J.Biol.Chem., 290, 2015
|
|
2E9Q
 
 | | Recombinant pro-11S globulin of pumpkin | | Descriptor: | 11S globulin subunit beta, CHLORIDE ION, PHOSPHATE ION | | Authors: | Fukuda, T, Prak, K, Itoh, T, Masuda, T, Maruyama, N, Mikami, B, Utsumi, S. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 2010
|
|
7CUG
 
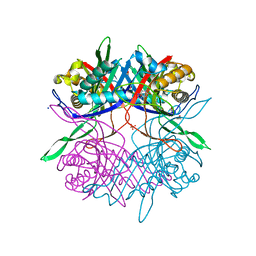 | |
2Z8R
 
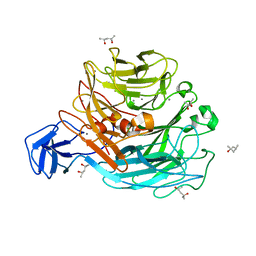 | | Crystal structure of rhamnogalacturonan lyase YesW at 1.40 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, YesW protein | | Authors: | Ochiai, A, Itoh, T, Maruyama, Y, Kawamata, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-09-10 | | Release date: | 2007-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Novel Structural Fold in Polysaccharide Lyases: BACILLUS SUBTILIS FAMILY 11 RHAMNOGALACTURONAN LYASE YesW WITH AN EIGHT-BLADED -PROPELLER
J.Biol.Chem., 282, 2007
|
|
2Z8S
 
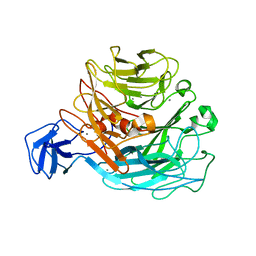 | | Crystal structure of rhamnogalacturonan lyase YesW complexed with digalacturonic acid | | Descriptor: | CALCIUM ION, YesW protein, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Ochiai, A, Itoh, T, Maruyama, Y, Kawamata, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-09-10 | | Release date: | 2007-10-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Structural Fold in Polysaccharide Lyases: BACILLUS SUBTILIS FAMILY 11 RHAMNOGALACTURONAN LYASE YesW WITH AN EIGHT-BLADED -PROPELLER
J.Biol.Chem., 282, 2007
|
|
2ZUX
 
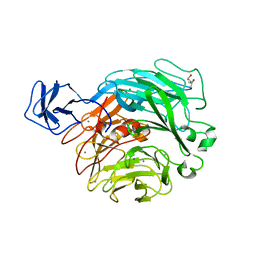 | | Crystal structure of rhamnogalacturonan lyase YesW complexed with rhamnose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, YesW protein, ... | | Authors: | Ochiai, A, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2008-10-28 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural determinants responsible for substrate recognition and mode of action in family 11 polysaccharide lyases
J.Biol.Chem., 284, 2009
|
|
