4UWM
 
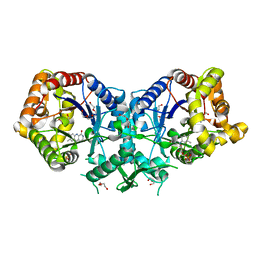 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2014-08-12 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7QYY
 
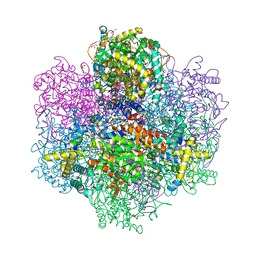 | | Vanadium-dependent bromoperoxidase from Corallina pilulifera in complex with chloride | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-29 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
1QHB
 
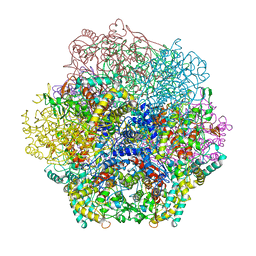 | | VANADIUM BROMOPEROXIDASE FROM RED ALGA CORALLINA OFFICINALIS | | Descriptor: | CALCIUM ION, HALOPEROXIDASE, PHOSPHATE ION | | Authors: | Isupov, M.N, Dalby, A.R, Brindley, A.A, Littlechild, J.A. | | Deposit date: | 1999-05-11 | | Release date: | 2000-07-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of dodecameric vanadium-dependent bromoperoxidase from the red algae Corallina officinalis.
J.Mol.Biol., 299, 2000
|
|
1QMV
 
 | | thioredoxin peroxidase B from red blood cells | | Descriptor: | PEROXIREDOXIN-2 | | Authors: | Isupov, M.N, Littlechild, J.A, Lebedev, A.A, Errington, N, Vagin, A.A, Schroder, E. | | Deposit date: | 1999-10-07 | | Release date: | 2000-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Decameric 2-Cys Peroxiredoxin from Human Erythrocytes at 1.7 A Resolution.
Structure, 8, 2000
|
|
1AX4
 
 | | TRYPTOPHANASE FROM PROTEUS VULGARIS | | Descriptor: | POTASSIUM ION, TRYPTOPHANASE | | Authors: | Isupov, M.N, Antson, A.A, Dodson, E.J, Dodson, G.G, Dementieva, I.S, Zakomirdina, L.N, Wilson, K.S, Dauter, Z, Lebedev, A.A, Harutyunyan, E.H. | | Deposit date: | 1997-10-28 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of tryptophanase.
J.Mol.Biol., 276, 1998
|
|
6YAK
 
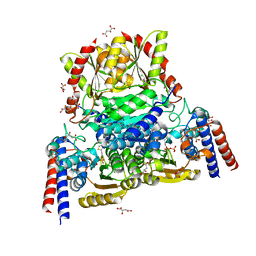 | | Split gene transketolase, active alpha2beta2 heterotetramer | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2H-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, C-terminal component of the split chain transketolase, ... | | Authors: | Isupov, M.N, Littlechild, J.A, James, P. | | Deposit date: | 2020-03-12 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A 'Split-Gene' Transketolase From the Hyper-Thermophilic Bacterium Carboxydothermus hydrogenoformans : Structure and Biochemical Characterization.
Front Microbiol, 11, 2020
|
|
6YAJ
 
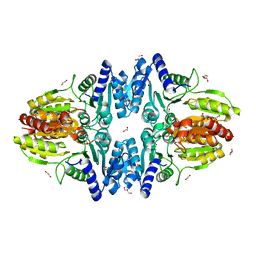 | | Split gene transketolase, inactive beta4 tetramer | | Descriptor: | 1,2-ETHANEDIOL, C-terminal chain of split transketolase from Carboxydothermus hydrogenoformans, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Littlechild, J.A, James, P. | | Deposit date: | 2020-03-12 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 'Split-Gene' Transketolase From the Hyper-Thermophilic Bacterium Carboxydothermus hydrogenoformans : Structure and Biochemical Characterization.
Front Microbiol, 11, 2020
|
|
6T92
 
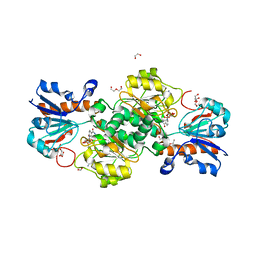 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex of N120C mutant protein with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T8Z
 
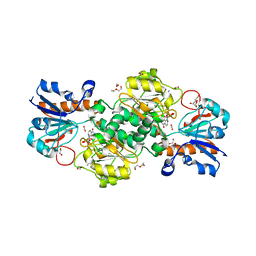 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A ternary complex with the oxidised form of the cofactor NAD+ and the substrate formate both at a primary and secondary sites. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T94
 
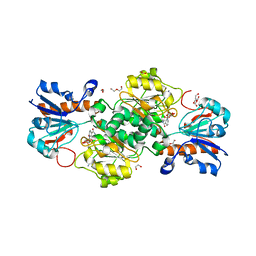 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex of N120C mutant protein with the reduced form of the cofactor NADH. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T8Y
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
1B7G
 
 | | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE | | Descriptor: | PROTEIN (GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE), SULFATE ION | | Authors: | Isupov, M.N, Littlechild, J.A. | | Deposit date: | 1999-01-22 | | Release date: | 1999-10-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus.
J.Mol.Biol., 291, 1999
|
|
1QGD
 
 | |
5MR0
 
 | | Thermophilic archaeal branched-chain amino acid transaminases from Geoglobus acetivorans and Archaeoglobus fulgidus: biochemical and structural characterisation | | Descriptor: | 1,2-ETHANEDIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, CHLORIDE ION, ... | | Authors: | Isupov, M.N, Littlechild, J.A, James, P, Sayer, C, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the ArchaeaGeoglobus acetivoransandArchaeoglobus fulgidus: Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
1XFF
 
 | | Glutaminase domain of glucosamine 6-phosphate synthase complexed with glutamate | | Descriptor: | ACETATE ION, GLUTAMIC ACID, Glucosamine--fructose-6-phosphate aminotransferase [isomerizing], ... | | Authors: | Isupov, M.N, Teplyakov, A. | | Deposit date: | 2004-09-14 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Binding is Required for Assembly of the Active Conformation of the Catalytic Site in Ntn Amidotransferases: Evidence from the 1.8 Angstrom Crystal Structure of the Glutaminase Domain of Glucosamine 6-Phosphate Synthase
Structure, 4, 1996
|
|
1XFG
 
 | | Glutaminase domain of glucosamine 6-phosphate synthase complexed with l-glu hydroxamate | | Descriptor: | ACETATE ION, GLUTAMINE HYDROXAMATE, Glucosamine--fructose-6-phosphate aminotransferase [isomerizing], ... | | Authors: | Isupov, M.N, Teplyakov, A. | | Deposit date: | 2004-09-14 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Binding is Required for Assembly of the Active Conformation of the Catalytic Site in Ntn Amidotransferases: Evidence from the 1.8 Angstrom Crystal Structure of the Glutaminase Domain of Glucosamine 6-Phosphate Synthase
Structure, 4, 1996
|
|
5AEC
 
 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2015-08-28 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7Q9X
 
 | | Crystal structure of Chromobacterium violaceum aminotransferase in complex with PLP-pyruvate adduct | | Descriptor: | (3E)-4-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}-2-oxobut-3-enoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Mitchell, D, Sayer, C, Littlechild, J.A. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aminotransferase from Chromobacterium violaceum in complex with PLP-pyruvate adduct.
To Be Published
|
|
7Q9Z
 
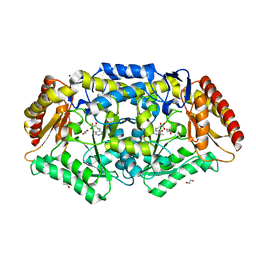 | | Crystal structure of Chromobacterium violaceum aminotransferase in complex with PLP-pyruvate adduct | | Descriptor: | (3E)-4-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}-2-oxobut-3-enoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Isupov, M.N, Mitchell, D, Sayer, C, Littlechild, J.A. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Aminotransferase from Chromobacterium violaceum in complex with PLP-pyruvate adduct.
To Be Published
|
|
7QW3
 
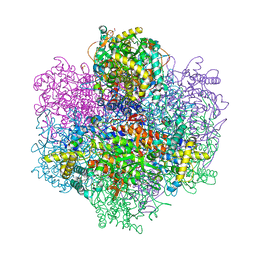 | | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera in complex with Br ion. | | Descriptor: | BROMIDE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-24 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
7QVW
 
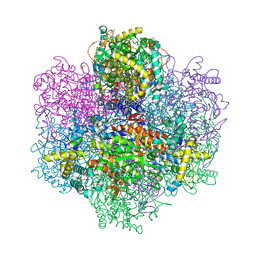 | | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera | | Descriptor: | CALCIUM ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
7QWI
 
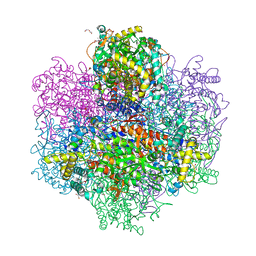 | | Vanadate complex of the vanadium-dependent bromoperoxidase from Corallina pilulifera | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CALCIUM ION, ... | | Authors: | Isupov, M.N, Mitchell, D, Littelchild, J.A, Garcia-Rodriguez, E. | | Deposit date: | 2022-01-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | R396W mutant of the vanadium-dependent bromoperoxidase from Corallina pilulifera
To Be Published
|
|
8QX4
 
 | | Sulfolobus acidocaldarius Archaellum filament. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-[alpha-D-mannopyranose-(1-4)][alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Flagellin, ... | | Authors: | Isupov, M.N, Gaines, M, McLaren, M, Daum, B. | | Deposit date: | 2023-10-22 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.03 Å) | | Cite: | Sulfolobus acidocaldarius Archaellum filament.
To Be Published
|
|
8RZL
 
 | | Sulfolobus acidocaldarius threads (0406) filament. | | Descriptor: | Sulfolobus acidocaldarius threads (0406) filament., alpha-D-mannopyranose, beta-D-glucopyranose-(1-4)-6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-[alpha-D-mannopyranose-(1-4)][alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Isupov, M.N, Gaines, M, McLaren, M, Daum, B. | | Deposit date: | 2024-02-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Sulfolobus acidocaldarius threads (0406) filament.
To Be Published
|
|
8AGS
 
 | | Cyclohexane epoxide soak of epoxide hydrolase from metagenomic source ch65 resulting in halogenated compound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 2-CHLOROPHENOL, Alpha/beta epoxide hydrolase, ... | | Authors: | Isupov, M.N, De Rose, S.A, Mitchell, D, Littlechild, J.A. | | Deposit date: | 2022-07-20 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Complexes of epoxide hydrolase from metagenomic source ch65
To Be Published
|
|
