8QPG
 
 | | Turret of Haloferax tailed virus 1 | | Descriptor: | MAGNESIUM ION, Prokaryotic polysaccharide deacetylase, ZINC ION, ... | | Authors: | Zhang, D, Daum, B, Isupov, M.N, McLaren, M. | | Deposit date: | 2023-10-02 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | CryoEM structure of Haloferax tailed virus 1
To Be Published
|
|
8QPQ
 
 | | C1 turret to capsid interface of full Haloferax tailed virus 1 adjacent to the portal-capsid interface. | | Descriptor: | HK97 gp5-like major capsid protein, MAGNESIUM ION, Prokaryotic polysaccharide deacetylase, ... | | Authors: | Zhang, D, Daum, B, Isupov, M.N, McLaren, M. | | Deposit date: | 2023-10-02 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | CryoEM structure of Haloferax tailed virus 1
To Be Published
|
|
8QQN
 
 | |
8QQJ
 
 | | CryoEM structure of the type IV pilin PilA5 from Thermus thermophilus | | Descriptor: | 7-Acetamido-5-acetimidoyl-3,5,7,9-tetradeoxy-L-glycero-L-manno-nonulosonic aci-(1-4)-alpha-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, MAGNESIUM ION, Type IV narrow pilus major component PilA5 | | Authors: | Gold, V.A.M, Neuhaus, A, Gaines, M, Isupov, M, McLaren, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | CryoEM structure of the type IV pilin PilA4 from Thermus thermophilus
To Be Published
|
|
8QSY
 
 | | Portal capsid interface of full Haloferax tailed virus 1. | | Descriptor: | Capsid stabilization protein, HK97 gp5-like major capsid protein, HK97 gp6-like/SPP1 gp15-like head-tail connector, ... | | Authors: | Zhang, D, Daum, B, Isupov, M.N, McLaren, M. | | Deposit date: | 2023-10-11 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | CryoEM structure of Haloferax tailed virus 1
To Be Published
|
|
8QQD
 
 | | CryoEM structure of the type IV pilin PilA4 from Thermus thermophilus | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, 7-Acetamido-5-acetimidoyl-3,5,7,9-tetradeoxy-L-glycero-L-manno-nonulosonic aci-(1-4)-alpha-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, ... | | Authors: | Gold, V.A.M, Neuhaus, A, Gaines, M, Isupov, M, McLaren, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | CryoEM structure of the type IV pilin PilA4 from Thermus thermophilus
To Be Published
|
|
8QSI
 
 | |
5NG7
 
 | | Novel epoxide hydrolases belonging to the alpha/beta hydrolases superfamily in metagenomes from hot environments | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ferrandi, E.E, De Rose, S.A, Sayer, C, Guazzelli, E, Marchesi, C, Saneei, V, Isupov, M.N, Littlechild, J.A, Monti, D. | | Deposit date: | 2017-03-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Thermophilic alpha / beta Class Epoxide Hydrolases Found in Metagenomes From Hot Environments.
Front Bioeng Biotechnol, 6, 2018
|
|
6J4R
 
 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*C)-3'), DNA (5'-D(*CP*CP*AP*TP*AP*TP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
6J5B
 
 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*GP*GP*TP*AP*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*CP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*TP*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*GP*TP*AP*CP*C)-3'), Protein PHOSPHATE STARVATION RESPONSE 1 | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N, Wu, Y.K. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
8PAJ
 
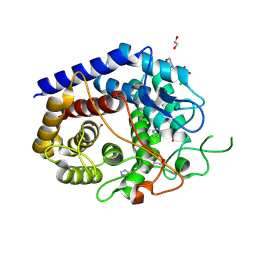 | | Crystal Structure of a Squalene-Hopene cyclase from Archangium gephyra | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Worthy, H.L, Isupov, M.N, Littlechild, J.A, Mitchell, D.E. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of a mesophilic Squalene-Hopene Cyclases from Cystobacter fuscus and Archangium gephyra
To Be Published
|
|
8PAK
 
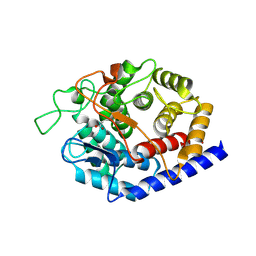 | | Crystal Structure of a Squalene-Hopene Cyclase from Cystobacter fuscus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Worthy, H.L, Mitchell, D.E, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure of Mesophilic Squalene-Hopene Cyclases from Cystobacter fuscus and Archangium gephyra
To Be Published
|
|
1T9H
 
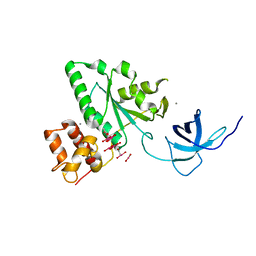 | | The crystal structure of YloQ, a circularly permuted GTPase. | | Descriptor: | ACETATE ION, CALCIUM ION, Probable GTPase engC, ... | | Authors: | Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Cladiere, L, Antson, A.A, Isupov, M.N, Seror, S.J, Wilkinson, A.J. | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of YloQ, a Circularly Permuted GTPase Essential for Bacillus Subtilis Viability.
J.Mol.Biol., 340, 2004
|
|
7PQ9
 
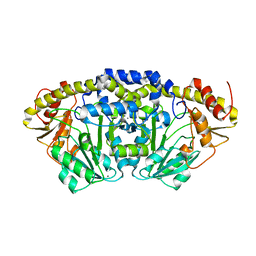 | | Crystal structure of Bacillus clausii pdxR at 2.8 Angstroms resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vivoli Vega, M, Isupov, M.N, Harmer, N. | | Deposit date: | 2021-09-16 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the DNA recognition mechanism by the bacterial transcription factor PdxR.
Nucleic Acids Res., 51, 2023
|
|
7PA1
 
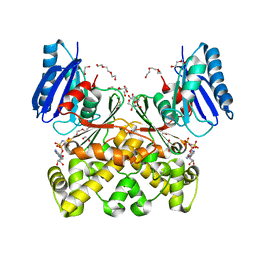 | | Structure of N-acetylglucosamine kinase from Plesiomonas shigelloides in complex with AMP-PNP in the absence of N-acetylglucoseamine substrate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7QCA
 
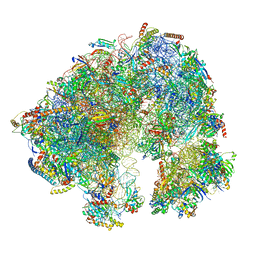 | | Spraguea lophii ribosome | | Descriptor: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S10, ... | | Authors: | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | Deposit date: | 2021-11-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state
Nat Microbiol, 2023
|
|
2WKW
 
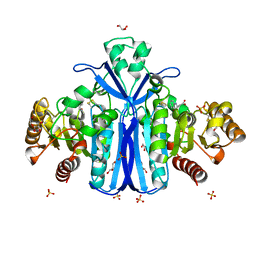 | | Alcaligenes esterase complexed with product analogue | | Descriptor: | CARBOXYLESTERASE, GLYCEROL, SULFATE ION, ... | | Authors: | Bourne, P.C, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2009-06-18 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Atomic-Resolution Structure of a Novel Bacterial Esterase.
Structure, 8, 2000
|
|
7ZCX
 
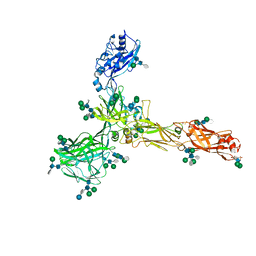 | | S-layer protein SlaA from Sulfolobus acidocaldarius at pH 4.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-deoxy-6-sulfo-beta-D-glucopyranose, ... | | Authors: | Gambelli, L, Isupov, M.N, Daum, B. | | Deposit date: | 2022-03-29 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the two-component S-layer of the archaeon Sulfolobus acidocaldarius.
Elife, 13, 2024
|
|
2W11
 
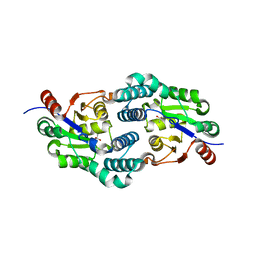 | | Structure of the L-2-haloacid dehalogenase from Sulfolobus tokodaii | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, 2-HALOALKANOIC ACID DEHALOGENASE | | Authors: | Rye, C.A, Isupov, M.N, Lebedev, A.A, Littlechild, J.A. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Structural Studies of a L-Haloacid Dehalogenase from the Thermophilic Archaeon Sulfolobus Tokodaii.
Extremophiles, 13, 2009
|
|
8AN3
 
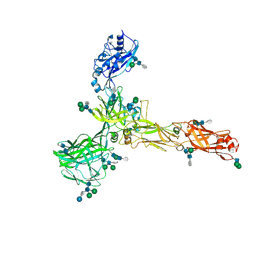 | | S-layer protein SlaA from Sulfolobus acidocaldarius at pH 7.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gambelli, L, Isupov, M.N, Daum, B. | | Deposit date: | 2022-08-04 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the two-component S-layer of the archaeon Sulfolobus acidocaldarius.
Elife, 13, 2024
|
|
8AN2
 
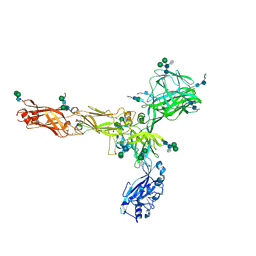 | | S-layer protein SlaA from Sulfolobus acidocaldarius at pH 10.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gambelli, L, Isupov, M.N, Daum, B. | | Deposit date: | 2022-08-04 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the two-component S-layer of the archaeon Sulfolobus acidocaldarius.
Elife, 13, 2024
|
|
8AXY
 
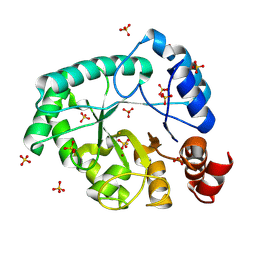 | | Staphylococcus aureus endonuclease IV orthorhombic crystal form | | Descriptor: | FE (III) ION, Probable endonuclease 4, SULFATE ION, ... | | Authors: | Rouvinski, A, Kirillov, S, Saper, M.A, Wiener, R, Isupov, M.N. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Octahedrally coordinated iron in the catalytic site of endonuclease IV from Staphylococcus aureus
To Be Published
|
|
7ZUG
 
 | | Heterogeneous nuclear ribonucleoprotein H1, qRRM2 domain | | Descriptor: | CHLORIDE ION, Heterogeneous nuclear ribonucleoprotein H, N-terminally processed, ... | | Authors: | Winter, N, Kumar, M, Isupov, M.N, Wiener, R. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.075 Å) | | Cite: | Heterogeneous nuclear ribonucleoprotein H1, qRRM2 domain
To Be Published
|
|
2WDU
 
 | | Fasciola hepatica sigma class GST | | Descriptor: | BROMIDE ION, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Line, K, Isupov, M.N, LaCourse, E.J, Brophy, P.M, Littlechild, J.A. | | Deposit date: | 2009-03-26 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The 1.6 Angstrom Crystal Structure of the Fasciola Hepatica Sigma Class Gst
To be Published
|
|
2WRT
 
 | | The 2.4 Angstrom structure of the Fasciola hepatica mu class GST, GST26 | | Descriptor: | CHLORIDE ION, GLUTATHIONE S-TRANSFERASE CLASS-MU 26 KDA ISOZYME 51 | | Authors: | Line, K, Isupov, M.N, LaCourse, E.J, Brophy, P.M, Littlechild, J.A. | | Deposit date: | 2009-09-02 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 2.5 Angstrom Structure of a Mu Class Gst from Fasciola Hepatica
To be Published
|
|
