7T1T
 
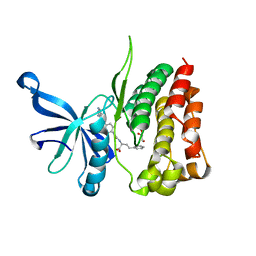 | | JAK2 JH2 IN COMPLEX WITH JAK292 | | Descriptor: | (2S)-2-[({4-[(2-amino-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy]phenyl}carbamoyl)amino]-4-phenylbutanoic acid, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Ippolito, J.A, Henry, S, Krimmer, S.G, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2021-12-02 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Conversion of a False Virtual Screen Hit into Selective JAK2 JH2 Domain Binders Using Convergent Design Strategies
Acs Med.Chem.Lett., 13, 2022
|
|
7T0P
 
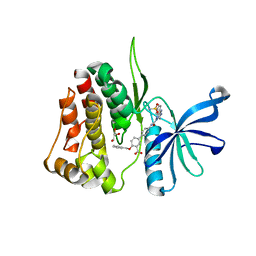 | | JAK2 JH2 IN COMPLEX WITH JAK315 | | Descriptor: | 4'-{[5-amino-3-(4-sulfamoylanilino)-1H-1,2,4-triazole-1-carbonyl]amino}-4-(benzyloxy)[1,1'-biphenyl]-3-carboxylic acid, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Ippolito, J.A, Liosi, M.-E, Krimmer, S.G, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2021-11-30 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Insights on JAK2 Modulation by Potent, Selective, and Cell-Permeable Pseudokinase-Domain Ligands.
J.Med.Chem., 65, 2022
|
|
1CCU
 
 | |
1CCT
 
 | |
1CCS
 
 | |
1CVD
 
 | |
1CVH
 
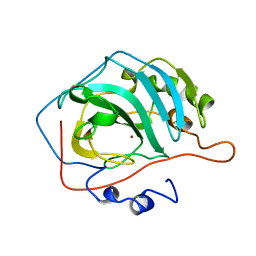 | |
1CNB
 
 | |
1CVE
 
 | |
397D
 
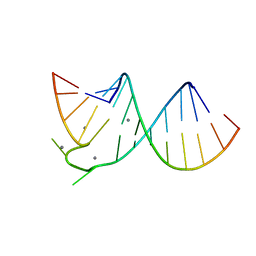 | |
3CPW
 
 | | The structure of the antibiotic LINEZOLID bound to the large ribosomal subunit of HALOARCULA MARISMORTUI | | Descriptor: | 23S RIBOSOMAL RNA, 5'-R(*CP*CP*AP*(PHE)*(ACA))-3', 50S ribosomal protein L10E, ... | | Authors: | Ippolito, J.A, Kanyo, Z.K, Wang, D, Franceschi, F.J, Moore, P.B, Steitz, T.A, Duffy, E.M. | | Deposit date: | 2008-04-01 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Oxazolidinone Antibiotic
Linezolid Bound to the 50S Ribosomal Subunit
J.Med.Chem., 51, 2008
|
|
3CXC
 
 | | The structure of an enhanced oxazolidinone inhibitor bound to the 50S ribosomal subunit of H. marismortui | | Descriptor: | (3Z)-N-[(4E)-5-(4-{(5S)-5-[(acetylamino)methyl]-2-oxo-1,3-oxazolidin-3-yl}-2-fluorophenyl)pent-4-en-1-yl]-3-(4-methyl-2,6-dioxo-1,6-dihydropyrimidin-5(2H)-ylidene)propanamide, 23S RIBOSOMAL RNA, 5'-R(*CP*CP*A)-3', ... | | Authors: | Ippolito, J.A, Wang, D, Kanyo, Z.F, Duffy, E.M. | | Deposit date: | 2008-04-24 | | Release date: | 2009-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design at the atomic level: design of biaryloxazolidinones as potent orally active antibiotics.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1CNC
 
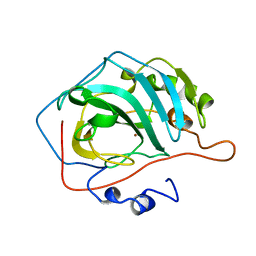 | |
1CVC
 
 | |
1CSL
 
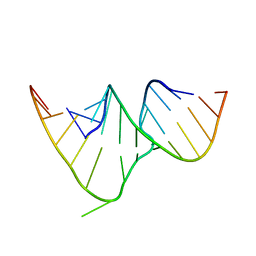 | | CRYSTAL STRUCTURE OF THE RRE HIGH AFFINITY SITE | | Descriptor: | 5'-R(*AP*AP*CP*GP*GP*GP*CP*GP*CP*AP*GP*AP*A)-3', 5'-R(*UP*CP*UP*GP*AP*CP*GP*GP*UP*AP*CP*GP*UP*UP*U)-3' | | Authors: | Ippolito, J.A, Steitz, T.A. | | Deposit date: | 1999-08-18 | | Release date: | 2000-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the HIV-1 RRE high affinity rev binding site at 1.6 A resolution.
J.Mol.Biol., 295, 2000
|
|
1CVB
 
 | |
1CVF
 
 | |
1CVA
 
 | |
1DCA
 
 | |
1DCB
 
 | |
7N44
 
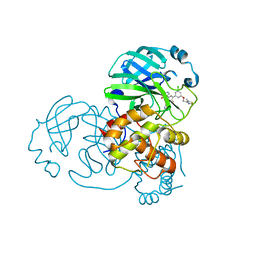 | | Crystal structure of the SARS-CoV-2 (2019-NCoV) main protease in complex with 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione (compound 13) | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Reilly, R.A, Zhang, C.H, Deshmukh, M.G, Ippolito, J.A, Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-06-03 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Optimization of Triarylpyridinone Inhibitors of the Main Protease of SARS-CoV-2 to Low-Nanomolar Antiviral Potency.
Acs Med.Chem.Lett., 12, 2021
|
|
7M8Y
 
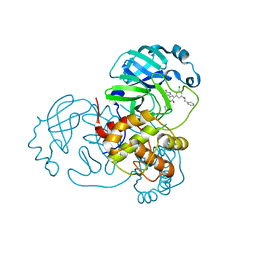 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 15 | | Descriptor: | 3C-like proteinase, 5-{3-[3-chloro-5-(2-phenylethoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M90
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 50 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[2-(3-oxopiperazin-1-yl)ethoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M8O
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 19 | | Descriptor: | 3C-like proteinase, 5-(3-{3-chloro-5-[(3-fluorophenyl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
7M91
 
 | | CRYSTAL STRUCTURE OF THE SARS-COV-2(2019-NCOV) MAIN PROTEASE IN COMPLEX WITH COMPOUND 25 | | Descriptor: | 3C-like proteinase, 5-{3-[3-chloro-5-(3,3,3-trifluoropropoxy)phenyl]-2-oxo-2H-[1,3'-bipyridin]-5-yl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Deshmukh, M.G, Ippolito, J.A, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease.
Structure, 29, 2021
|
|
