4GHL
 
 | | Structural Basis for Marburg virus VP35 mediate immune evasion mechanisms | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Ramanan, P, Borek, D.M, Otwinowski, Z, Leung, D.W, Amarasinghe, G.K. | | 登録日 | 2012-08-07 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural basis for Marburg virus VP35-mediated immune evasion mechanisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1AOY
 
 | |
3GRX
 
 | | NMR STRUCTURE OF ESCHERICHIA COLI GLUTAREDOXIN 3-GLUTATHIONE MIXED DISULFIDE COMPLEX, 20 STRUCTURES | | 分子名称: | GLUTAREDOXIN 3, GLUTATHIONE | | 著者 | Nordstrand, K, Aslund, F, Holmgren, A, Otting, G, Berndt, K.D. | | 登録日 | 1998-08-17 | | 公開日 | 1999-03-30 | | 最終更新日 | 2018-03-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of Escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: implications for the enzymatic mechanism.
J.Mol.Biol., 286, 1999
|
|
1ZFO
 
 | | AMINO-TERMINAL LIM-DOMAIN PEPTIDE OF LASP-1, NMR | | 分子名称: | LASP-1, ZINC ION | | 著者 | Hammarstrom, A, Berndt, K.D, Sillard, R, Adermann, K, Otting, G. | | 登録日 | 1996-05-06 | | 公開日 | 1996-11-08 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a naturally-occurring zinc-peptide complex demonstrates that the N-terminal zinc-binding module of the Lasp-1 LIM domain is an independent folding unit.
Biochemistry, 35, 1996
|
|
2ARR
 
 | | Human plasminogen activator inhibitor-2.[loop (66-98) deletion mutant] complexed with peptide n-acetyl-teaaagmggvmtgr-oh | | 分子名称: | 14-mer from Plasminogen activator inhibitor-2, Plasminogen activator inhibitor-2 | | 著者 | Di Giusto, D.A, Sutherland, A.P, Jankova, L, Harrop, S.J, Curmi, P.M, King, G.C. | | 登録日 | 2005-08-21 | | 公開日 | 2006-07-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Plasminogen activator inhibitor-2 is highly tolerant to P8 residue substitution--implications for serpin mechanistic model and prediction of nsSNP activities
J.Mol.Biol., 353, 2005
|
|
1HP3
 
 | |
3Q5U
 
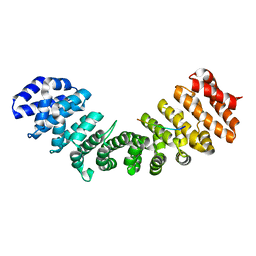 | |
1CCD
 
 | | REFINED STRUCTURE OF RAT CLARA CELL 17 KDA PROTEIN AT 3.0 ANGSTROMS RESOLUTION | | 分子名称: | CLARA CELL 17 kD PROTEIN, SULFATE ION | | 著者 | Umland, T.C, Swaminathan, S, Furey, W, Singh, G, Pletcher, J, Sax, M. | | 登録日 | 1991-09-17 | | 公開日 | 1994-01-31 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Refined structure of rat Clara cell 17 kDa protein at 3.0 A resolution.
J.Mol.Biol., 224, 1992
|
|
4PP7
 
 | | Highly Potent and Selective 3-N-methylquinazoline-4(3H)-one Based Inhibitors of B-RafV600E Kinase | | 分子名称: | N-{2,4-difluoro-3-[methyl(3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]phenyl}propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | 著者 | Wenglowsky, S, Ren, L, Grina, J, Hansen, J.D, Laird, E.R, Moreno, D, Dinkel, V, Gloor, S.L, Hastings, G, Rana, S, Rasor, K, Sturgis, H.L, Voegtli, W.C, Vigers, G.P.A, Willis, B, Mathieu, S, Rudolph, J. | | 登録日 | 2014-02-26 | | 公開日 | 2014-04-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Highly potent and selective 3-N-methylquinazoline-4(3H)-one based inhibitors of B-Raf(V600E) kinase.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1AXJ
 
 | |
3FYR
 
 | | Crystal structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis | | 分子名称: | Sporulation inhibitor sda | | 著者 | Jacques, D.A, Streamer, M, King, G.F, Guss, J.M, Trewhella, J, Langley, D.B. | | 登録日 | 2009-01-23 | | 公開日 | 2009-06-23 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis and insights into its solution state
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1MSZ
 
 | | Solution structure of the R3H domain from human Smubp-2 | | 分子名称: | DNA-binding protein SMUBP-2 | | 著者 | Liepinsh, E, Leonchiks, A, Sharipo, A, Guignard, L, Otting, G. | | 登録日 | 2002-09-20 | | 公開日 | 2002-10-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the R3H domain from human Smubp-2
J.Mol.Biol., 326, 2003
|
|
2YBA
 
 | | Crystal structure of Nurf55 in complex with histone H3 | | 分子名称: | HISTONE H3, PROBABLE HISTONE-BINDING PROTEIN CAF1 | | 著者 | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | 登録日 | 2011-03-02 | | 公開日 | 2011-05-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks
Mol.Cell, 42, 2011
|
|
2YB8
 
 | | Crystal structure of Nurf55 in complex with Su(z)12 | | 分子名称: | POLYCOMB PROTEIN SU(Z)12, PROBABLE HISTONE-BINDING PROTEIN CAF1, SULFATE ION | | 著者 | Schmitges, F.W, Prusty, A.B, Faty, M, Stutzer, A, Lingaraju, G.M, Aiwazian, J, Sack, R, Hess, D, Li, L, Zhou, S, Bunker, R.D, Wirth, U, Bouwmeester, T, Bauer, A, Ly-Hartig, N, Zhao, K, Chan, H, Gu, J, Gut, H, Fischle, W, Muller, J, Thoma, N.H. | | 登録日 | 2011-03-02 | | 公開日 | 2011-05-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Histone Methylation by Prc2 is Inhibited by Active Chromatin Marks.
Mol.Cell, 42, 2011
|
|
7YLA
 
 | | Cryo-EM structure of 50S-HflX complex | | 分子名称: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Damu, W, Ning, G. | | 登録日 | 2022-07-25 | | 公開日 | 2023-01-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (2.52 Å) | | 主引用文献 | Cryo-EM Structure of the 50S-HflX Complex Reveals a Novel Mechanism of Antibiotic Resistance in E. coli
Biorxiv, 2022
|
|
4EI2
 
 | |
1HVW
 
 | |
7YQK
 
 | | cryo-EM structure of gammaH2AXK15ub-H4K20me2 nucleosome bound to 53BP1 | | 分子名称: | DNA (145-MER), Histone H2AX, Histone H2B, ... | | 著者 | Ai, H.S, GuoChao, C, Qingyue, G, Ze-Bin, T, Zhiheng, D, Xin, L, Fan, Y, Ziyu, X, Jia-Bin, L, Changlin, T, Liu, L. | | 登録日 | 2022-08-07 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | Chemical Synthesis of Post-Translationally Modified H2AX Reveals Redundancy in Interplay between Histone Phosphorylation, Ubiquitination, and Methylation on the Binding of 53BP1 with Nucleosomes.
J.Am.Chem.Soc., 144, 2022
|
|
3UBY
 
 | | Crystal structure of human alklyadenine DNA glycosylase in a lower and higher-affinity complex with DNA | | 分子名称: | DNA (5'-D(*GP*AP*CP*AP*TP*GP*(EDC)P*TP*TP*GP*CP*CP*T)-3'), DNA-3-methyladenine glycosylase | | 著者 | Setser, J.W, Lingaraju, G.M, Davis, C.A, Samson, L.D, Drennan, C.L. | | 登録日 | 2011-10-25 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Searching for DNA lesions: structural evidence for lower- and higher-affinity DNA binding conformations of human alkyladenine DNA glycosylase.
Biochemistry, 51, 2012
|
|
5TBK
 
 | | Crystal structure of human importin a3 bound to RCC1 | | 分子名称: | Importin subunit alpha-3, Regulator of chromosome condensation | | 著者 | Sankhala, R.S, Lokareddy, R.K, Pumroy, R.A, Cingolani, G. | | 登録日 | 2016-09-12 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Three-dimensional context rather than NLS amino acid sequence determines importin alpha subtype specificity for RCC1.
Nat Commun, 8, 2017
|
|
2AYA
 
 | |
3DA7
 
 | | A conformationally strained, circular permutant of barnase | | 分子名称: | Barnase circular permutant, Barstar | | 著者 | Mitrousis, G, Butler, J, Loh, S.N, Cingolani, G. | | 登録日 | 2008-05-28 | | 公開日 | 2009-04-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural and thermodynamic analysis of a conformationally strained circular permutant of barnase.
Biochemistry, 48, 2009
|
|
1WNJ
 
 | | NMR structure of human coactosin-like protein | | 分子名称: | Coactosin-like protein | | 著者 | Liepinsh, E, Rakonjac, M, Boissonneault, V, Provost, P, Samuelsson, B, Radmark, O, Otting, G. | | 登録日 | 2004-08-05 | | 公開日 | 2004-08-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of human coactosin-like protein
J.Biomol.Nmr, 30, 2004
|
|
3TJ3
 
 | | Structure of importin a5 bound to the N-terminus of Nup50 | | 分子名称: | Importin subunit alpha-1, Nuclear pore complex protein Nup50 | | 著者 | Pumroy, R, Nardozzi, J.D, Hart, D.J, Root, M.J, Cingolani, G. | | 登録日 | 2011-08-23 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.702 Å) | | 主引用文献 | Nucleoporin Nup50 stabilizes closed conformation of armadillo repeat 10 in importin alpha 5.
J.Biol.Chem., 287, 2012
|
|
1FOV
 
 | | GLUTAREDOXIN 3 FROM ESCHERICHIA COLI IN THE FULLY OXIDIZED FORM | | 分子名称: | GLUTAREDOXIN 3 | | 著者 | Nordstrand, K, Sandstrom, A, Aslund, F, Holmgren, A, Otting, G, Berndt, K.D. | | 登録日 | 2000-08-29 | | 公開日 | 2000-10-26 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of oxidized glutaredoxin 3 from Escherichia coli.
J.Mol.Biol., 303, 2000
|
|
