6A0J
 
 | | Cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from Arthrobacter globiformis, complex with Cyclic alpha-maltosyl-(1-->6)-maltose | | Descriptor: | CALCIUM ION, Cyclic alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Cyclic maltosyl-maltose hydrolase | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural features of a bacterial cyclic alpha-maltosyl-(1→6)-maltose (CMM) hydrolase critical for CMM recognition and hydrolysis.
J. Biol. Chem., 293, 2018
|
|
6A0K
 
 | | Cyclic alpha-maltosyl-(1-->6)-maltose hydrolase from Arthrobacter globiformis, complex with panose | | Descriptor: | CALCIUM ION, Cyclic maltosyl-maltose hydrolase, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural features of a bacterial cyclic alpha-maltosyl-(1→6)-maltose (CMM) hydrolase critical for CMM recognition and hydrolysis.
J. Biol. Chem., 293, 2018
|
|
1WM1
 
 | | Crystal Structure of Prolyl Aminopeptidase, Complex with Pro-TBODA | | Descriptor: | (5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)[(2R)-PYRROLIDIN-2-YL]METHANONE, Proline iminopeptidase | | Authors: | Nakajima, Y, Inoue, T, Ito, K, Tozaka, T, Hatakeyama, S, Tanaka, N, Nakamura, K.T, Yoshimoto, T. | | Deposit date: | 2004-07-01 | | Release date: | 2004-07-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel inhibitor for prolyl aminopeptidase from Serratia marcescens and studies on the mechanism of substrate recognition of the enzyme using the inhibitor
ARCH.BIOCHEM.BIOPHYS., 416, 2003
|
|
1WMZ
 
 | | Crystal Structure of C-type Lectin CEL-I complexed with N-acetyl-D-galactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Sugawara, H, Kusunoki, M, Kurisu, G, Fujimoto, T, Aoyagi, H, Hatakeyama, T. | | Deposit date: | 2004-07-22 | | Release date: | 2004-09-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characteristic Recognition of N-Acetylgalactosamine by an Invertebrate C-type Lectin, CEL-I, Revealed by X-ray Crystallographic Analysis
J.Biol.Chem., 279, 2004
|
|
5Y90
 
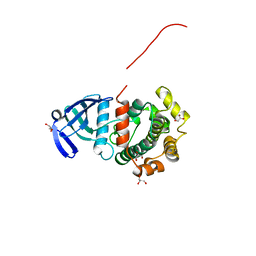 | | MAP2K7 mutant -C218S | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7, GLYCEROL | | Authors: | Kinoshita, T, Hashimoto, T, Sogabe, Y, Matsumoto, T, Sawa, M, Fukada, H. | | Deposit date: | 2017-08-22 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structure discloses the potential for allosteric regulation of mitogen-activated protein kinase kinase 7
Biochem. Biophys. Res. Commun., 493, 2017
|
|
1RRP
 
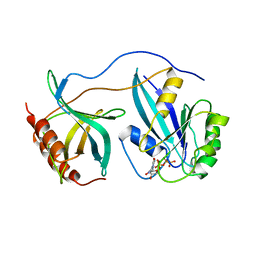 | | STRUCTURE OF THE RAN-GPPNHP-RANBD1 COMPLEX | | Descriptor: | MAGNESIUM ION, NUCLEAR PORE COMPLEX PROTEIN NUP358, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Vetter, I.R, Nowak, C, Nishimoto, T, Kuhlmann, J, Wittinghofer, A. | | Deposit date: | 1999-01-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport.
Nature, 398, 1999
|
|
2AE2
 
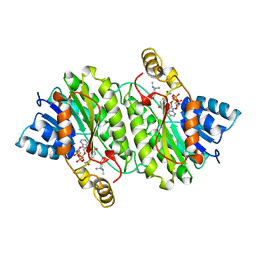 | | TROPINONE REDUCTASE-II COMPLEXED WITH NADP+ AND PSEUDOTROPINE | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (TROPINONE REDUCTASE-II), PSEUDOTROPINE | | Authors: | Yamashita, A, Kato, H, Wakatsuki, S, Tomizaki, T, Nakatsu, T, Nakajima, K, Hashimoto, T, Yamada, Y, Oda, J. | | Deposit date: | 1999-01-26 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of tropinone reductase-II complexed with NADP+ and pseudotropine at 1.9 A resolution: implication for stereospecific substrate binding and catalysis.
Biochemistry, 38, 1999
|
|
1WMY
 
 | | Crystal Structure of C-type Lectin CEL-I from Cucumaria echinata | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, lectin CEL-I, ... | | Authors: | Sugawara, H, Kusunoki, M, Kurisu, G, Fujimoto, T, Aoyagi, H, Hatakeyama, T. | | Deposit date: | 2004-07-22 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characteristic Recognition of N-Acetylgalactosamine by an Invertebrate C-type Lectin, CEL-I, Revealed by X-ray Crystallographic Analysis
J.Biol.Chem., 279, 2004
|
|
1UKL
 
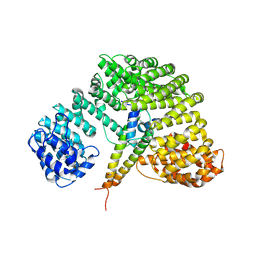 | | Crystal structure of Importin-beta and SREBP-2 complex | | Descriptor: | Importin beta-1 subunit, Sterol regulatory element binding protein-2 | | Authors: | Lee, S.J, Sekimoto, T, Yamashita, E, Nagoshi, E, Nakagawa, A, Imamoto, N, Yoshimura, M, Sakai, H, Tsukihara, T, Yoneda, Y. | | Deposit date: | 2003-08-26 | | Release date: | 2003-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Importin-beta Bound to SREBP-2: Nuclear Import of a Transcription Factor
Science, 302, 2003
|
|
5YJB
 
 | | LSD1-CoREST in complex with 4-[5-(piperidin-4-ylmethoxy)-2-(p-tolyl)pyridin-3-yl]benzonitrile | | Descriptor: | 4-[2-(4-methylphenyl)-5-(piperidin-4-ylmethoxy)pyridin-3-yl]benzenecarbonitrile, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Hashimoto, T, Matsuno, K, Umehara, T. | | Deposit date: | 2017-10-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal Structure of LSD1 in Complex with 4-[5-(Piperidin-4-ylmethoxy)-2-(p-tolyl)pyridin-3-yl]benzonitrile.
Molecules, 23, 2018
|
|
2AE1
 
 | | TROPINONE REDUCTASE-II | | Descriptor: | TROPINONE REDUCTASE-II | | Authors: | Nakajima, K, Yamashita, A, Akama, H, Nakatsu, T, Kato, H, Hashimoto, T, Oda, J, Yamada, Y. | | Deposit date: | 1997-10-27 | | Release date: | 1998-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of two tropinone reductases: different reaction stereospecificities in the same protein fold.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1KOL
 
 | | Crystal structure of formaldehyde dehydrogenase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ZINC ION, ... | | Authors: | Tanaka, N, Kusakabe, Y, Ito, K, Yoshimoto, T, Nakamura, K.T. | | Deposit date: | 2001-12-21 | | Release date: | 2002-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Formaldehyde Dehydrogenase from Pseudomonas putida: the Structural Origin of the Tightly Bound Cofactor in Nicotinoprotein Dehydrogenases
J.mol.biol., 324, 2002
|
|
1E8T
 
 | | Structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Crennell, S, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2000-10-01 | | Release date: | 2001-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Multifunctional Paramyxovirus Hemagglutinin-Neuraminidase
Nat.Struct.Biol., 7, 2000
|
|
3WIQ
 
 | | Crystal structure of kojibiose phosphorylase complexed with kojibiose | | Descriptor: | Kojibiose phosphorylase, SULFATE ION, alpha-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Okada, S, Yamamoto, T, Watanabe, H, Nishimoto, T, Chaen, H, Fukuda, S, Wakagi, T, Fushinobu, S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and mutational analysis of substrate recognition in kojibiose phosphorylase
Febs J., 281, 2014
|
|
3WIR
 
 | | Crystal structure of kojibiose phosphorylase complexed with glucose | | Descriptor: | GLYCEROL, Kojibiose phosphorylase, PHOSPHATE ION, ... | | Authors: | Okada, S, Yamamoto, T, Watanabe, H, Nishimoto, T, Chaen, H, Fukuda, S, Wakagi, T, Fushinobu, S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and mutational analysis of substrate recognition in kojibiose phosphorylase
Febs J., 281, 2014
|
|
1USX
 
 | | Crystal structure of the Newcastle disease virus hemagglutinin-neuraminidase complexed with thiosialoside | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, HEMAGGLUTININ-NEURAMINIDASE GLYCOPROTEIN, N-acetyl-alpha-neuraminic acid-(2-6)-methyl 6-thio-beta-D-galactopyranoside | | Authors: | Zaitsev, V, Itzstein, M, Groves, D, Kiefel, M, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Second Sialic Acid Binding Site in Newcastle Disease Virus Hemagglutinin-Neuraminidase: Implications for Fusion
J.Virol., 78, 2004
|
|
1USR
 
 | | Newcastle disease virus hemagglutinin-neuraminidase: Evidence for a second sialic acid binding site and implications for fusion | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zaitsev, V, Von Itzstein, M, Groves, D, Kiefel, M, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2003-11-28 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Second Sialic Acid Binding Site in Newcastle Disease Virus Hemagglutinin-Neuraminidase: Implications for Fusion
J.Virol., 78, 2004
|
|
1E8V
 
 | | Structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Crennell, S, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2000-10-01 | | Release date: | 2001-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Multifunctional Paramyxovirus Hemagglutinin-Neuraminidase
Nat.Struct.Biol., 7, 2000
|
|
1E8U
 
 | | Structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HEMAGGLUTININ-NEURAMINIDASE, ... | | Authors: | Crennell, S, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2000-10-01 | | Release date: | 2001-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Multifunctional Paramyxovirus Hemagglutinin-Neuraminidase
Nat.Struct.Biol., 7, 2000
|
|
2DCM
 
 | | The Crystal Structure of S603A Mutated Prolyl Tripeptidyl Aminopeptidase Complexed with Substrate | | Descriptor: | GLYCYLALANYL-N-2-NAPHTHYL-L-PROLINEAMIDE, dipeptidyl aminopeptidase IV, putative | | Authors: | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | Deposit date: | 2006-01-09 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
2EEP
 
 | | Prolyl Tripeptidyl Aminopeptidase Complexed with an Inhibitor | | Descriptor: | Dipeptidyl aminopeptidase IV, putative, SULFATE ION, ... | | Authors: | Xu, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2007-02-16 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel inhibitor for prolyl tripeptidyl aminopeptidase from Porphyromonas gingivalis and details of substrate-recognition mechanism
J.Mol.Biol., 375, 2008
|
|
2ECF
 
 | |
2D5L
 
 | | Crystal Structure of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis | | Descriptor: | SULFATE ION, dipeptidyl aminopeptidase IV, putative | | Authors: | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | Deposit date: | 2005-11-02 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
2DQM
 
 | | Crystal Structure of Aminopeptidase N complexed with bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Aminopeptidase N, SULFATE ION, ... | | Authors: | Onohara, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2006-05-29 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
2DQ6
 
 | | Crystal Structure of Aminopeptidase N from Escherichia coli | | Descriptor: | Aminopeptidase N, SULFATE ION, ZINC ION | | Authors: | Nakajima, Y, Onohara, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2006-05-22 | | Release date: | 2006-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
