1CCZ
 
 | | CRYSTAL STRUCTURE OF THE CD2-BINDING DOMAIN OF CD58 (LYMPHOCYTE FUNCTION-ASSOCIATED ANTIGEN 3) AT 1.8-A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CD58) | | Authors: | Ikemizu, S, Sparks, L.M, Van Der Merwe, P.A, Harlos, K, Stuart, D.I, Jones, E.Y, Davis, S.J. | | Deposit date: | 1999-03-02 | | Release date: | 1999-04-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the CD2-binding domain of CD58 (lymphocyte function-associated antigen 3) at 1.8-A resolution.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1DR9
 
 | | CRYSTAL STRUCTURE OF A SOLUBLE FORM OF B7-1 (CD80) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, T LYMPHOCYTE ACTIVATION ANTIGEN | | Authors: | Ikemizu, S, Jones, E.Y, Stuart, D.I, Davis, S.J. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and dimerization of a soluble form of B7-1.
Immunity, 12, 2000
|
|
3NG5
 
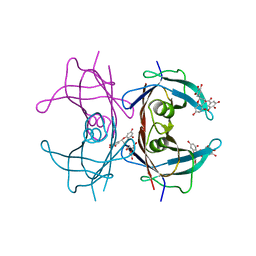 | | Crystal Structure of V30M transthyretin complexed with (-)-epigallocatechin gallate (EGCG) | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, GLYCEROL, Transthyretin | | Authors: | Miyata, M, Nakamura, T, Ikemizu, S, Chirifu, M, Yamagata, Y, Kai, H. | | Deposit date: | 2010-06-11 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of green tea polyphenol(-)-epigallocatechin gallate (EGCG)-transthyretin complex reveals novel binding site distinct from thyroxine binding site
Biochemistry, 2010
|
|
2M2D
 
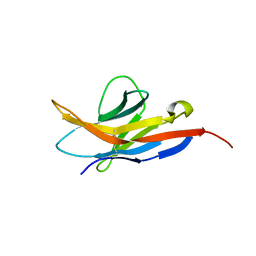 | | Human programmed cell death 1 receptor | | Descriptor: | Programmed cell death protein 1 | | Authors: | Veverka, V, Cheng, X, Waters, L.C, Muskett, F.W, Morgan, S, Lesley, A, Griffiths, M, Stubberfield, C, Griffin, R, Henry, A.J, Robinson, M.K, Jansson, A, Ladbury, J.E, Ikemizu, S, Davis, S.J, Carr, M.D. | | Deposit date: | 2012-12-18 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of the human programmed cell death 1 receptor.
J.Biol.Chem., 288, 2013
|
|
3OSK
 
 | | Crystal structure of human CTLA-4 apo homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytotoxic T-lymphocyte protein 4, GLYCEROL | | Authors: | Yu, C, Sonnen, A.F.-P, Ikemizu, S, Stuart, D.I, Gilbert, R.J.C, Davis, S.J. | | Deposit date: | 2010-09-09 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rigid-body ligand recognition drives cytotoxic T-lymphocyte antigen 4 (CTLA-4) receptor triggering
J.Biol.Chem., 286, 2011
|
|
2DRU
 
 | | Crystal structure and binding properties of the CD2 and CD244 (2B4) binding protein, CD48 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, chimera of CD48 antigen and T-cell surface antigen CD2 | | Authors: | Evans, E.J, Ikemizu, S, Davis, S.J. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Binding Properties of the CD2 and CD244 (2B4)-binding Protein, CD48
J.Biol.Chem., 281, 2006
|
|
2Z3Q
 
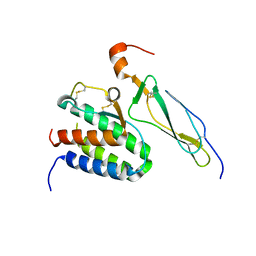 | | Crystal structure of the IL-15/IL-15Ra complex | | Descriptor: | Interleukin-15, Interleukin-15 receptor alpha chain | | Authors: | Chirifu, M, Yamagata, Y, Davis, S.J, Ikemizu, S. | | Deposit date: | 2007-06-05 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the IL-15-IL-15Ralpha complex, a cytokine-receptor unit presented in trans
Nat.Immunol., 8, 2007
|
|
3A4F
 
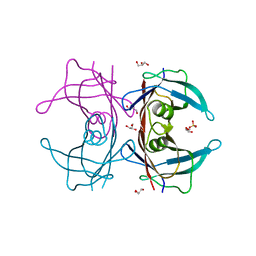 | | Crystal Structure of Human Transthyretin (E54K) | | Descriptor: | GLYCEROL, SULFATE ION, Transthyretin | | Authors: | Miyata, M, Sato, T, Nakamura, T, Ikemizu, S, Yamagata, Y, Kai, H. | | Deposit date: | 2009-07-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Role of the glutamic acid 54 residue in transthyretin stability and thyroxine binding
Biochemistry, 49, 2010
|
|
2Z3R
 
 | | Crystal structure of the IL-15/IL-15Ra complex | | Descriptor: | GLYCEROL, Interleukin-15, Interleukin-15 receptor alpha chain | | Authors: | Chirifu, M, Yamagata, Y, Davis, S.J, Ikemizu, S. | | Deposit date: | 2007-06-05 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the IL-15-IL-15Ralpha complex, a cytokine-receptor unit presented in trans
Nat.Immunol., 8, 2007
|
|
3A4E
 
 | | Crystal structure of Human Transthyretin (E54G) | | Descriptor: | GLYCEROL, SULFATE ION, Transthyretin | | Authors: | Miyata, M, Sato, T, Nakamura, T, Ikemizu, S, Yamagata, Y, Kai, H. | | Deposit date: | 2009-07-06 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of the glutamic acid 54 residue in transthyretin stability and thyroxine binding
Biochemistry, 49, 2010
|
|
1EVS
 
 | | CRYSTAL STRUCTURE OF HUMAN ONCOSTATIN M | | Descriptor: | ONCOSTATIN M | | Authors: | Deller, M.C, Hudson, K.R, Ikemizu, S, Bravo, J, Jones, E.Y, Heath, J.K. | | Deposit date: | 2000-04-20 | | Release date: | 2000-09-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and functional dissection of the cytostatic cytokine oncostatin M.
Structure Fold.Des., 8, 2000
|
|
1DTQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-1 (PETT131A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-NITRILOMETHYL-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-20 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1DTT
 
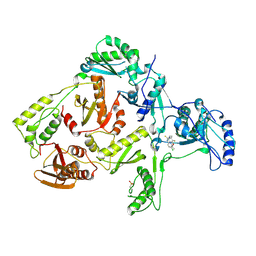 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 (PETT130A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-04-02 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1EPF
 
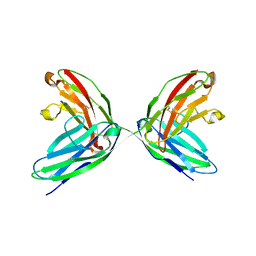 | | CRYSTAL STRUCTURE OF THE TWO N-TERMINAL IMMUNOGLOBULIN DOMAINS OF THE NEURAL CELL ADHESION MOLECULE (NCAM) | | Descriptor: | CALCIUM ION, PROTEIN (NEURAL CELL ADHESION MOLECULE) | | Authors: | Kasper, C, Rasmussen, H, Kastrup, J.S, Ikemizu, S, Jones, E.Y, Berezin, V, Bock, E, Larsen, I.K. | | Deposit date: | 2000-03-29 | | Release date: | 2000-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of cell-cell adhesion by NCAM.
Nat.Struct.Biol., 7, 2000
|
|
1H15
 
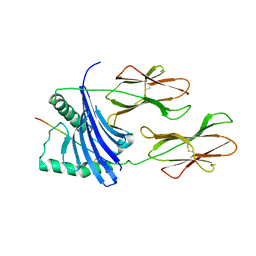 | | X-ray crystal structure of HLA-DRA1*0101/DRB5*0101 complexed with a peptide from Epstein Barr Virus DNA polymerase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA POLYMERASE, ... | | Authors: | Lang, H, Jacobsen, H, Ikemizu, S, Andersson, C, Harlos, K, Madsen, L, Hjorth, P, Sondergaard, L, Svejgaard, A, Wucherpfennig, K, Stuart, D.I, Bell, J.I, Jones, E.Y, Fugger, L. | | Deposit date: | 2002-07-02 | | Release date: | 2002-10-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Functional and Structural Basis for Tcr Cross-Reactivity in Multiple Sclerosis
Nat.Immunol., 3, 2002
|
|
