8S4T
 
 | |
8S4S
 
 | | PrgE from plasmid pCF10 | | Descriptor: | POTASSIUM ION, PrgE, SULFATE ION | | Authors: | Breidenstein, A, Berntsson, R.P.-A. | | Deposit date: | 2024-02-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | PrgE: an OB-fold protein from plasmid pCF10 with striking differences to prototypical bacterial SSBs.
Life Sci Alliance, 7, 2024
|
|
5VG9
 
 | | Structure of the eukaryotic intramembrane Ras methyltransferase ICMT (isoprenylcysteine carboxyl methyltransferase) without a monobody | | Descriptor: | Protein-S-isoprenylcysteine O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Long, S.B, Diver, M.M, Pedi, L, Koide, A, Koide, S. | | Deposit date: | 2017-04-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Atomic structure of the eukaryotic intramembrane RAS methyltransferase ICMT.
Nature, 553, 2018
|
|
2ZOE
 
 | | HA3 subcomponent of Clostridium botulinum type C progenitor toxin, complex with N-acetylneuramic acid | | Descriptor: | Hemagglutinin components HA3, N-acetyl-beta-neuraminic acid | | Authors: | Nakamura, T, Kotani, M, Tonozuka, T, Ide, A, Oguma, K, Nishikawa, A. | | Deposit date: | 2008-05-09 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the HA3 Subcomponent of Clostridium botulinum Type C Progenitor Toxin
J.Mol.Biol., 385, 2009
|
|
3AH4
 
 | | HA1 subcomponent of botulinum type C progenitor toxin complexed with galactose | | Descriptor: | Main hemagglutinin component, beta-D-galactopyranose | | Authors: | Nakamura, T, Tonozuka, T, Ide, A, Yuzawa, T, Oguma, K, Nishikawa, A. | | Deposit date: | 2010-04-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin
J.Mol.Biol., 376, 2008
|
|
3AH2
 
 | | HA1 subcomponent of botulinum type C progenitor toxin complexed with N-acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Main hemagglutinin component | | Authors: | Nakamura, T, Tonozuka, T, Ide, A, Yuzawa, T, Oguma, K, Nishikawa, A. | | Deposit date: | 2010-04-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin
J.Mol.Biol., 376, 2008
|
|
3AH1
 
 | | HA1 subcomponent of botulinum type C progenitor toxin complexed with N-acetylneuramic acid | | Descriptor: | Main hemagglutinin component, N-acetyl-beta-neuraminic acid | | Authors: | Nakamura, T, Tonozuka, T, Ide, A, Yuzawa, T, Oguma, K, Nishikawa, A. | | Deposit date: | 2010-04-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin
J.Mol.Biol., 376, 2008
|
|
6WW5
 
 | | Structure of VcINDY-Na-Fab84 in nanodisc | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, DASS family sodium-coupled anion symporter, Fab84 Heavy Chain, ... | | Authors: | Sauer, D.B, Marden, J, Song, J.M, Koide, A, Koide, S, Wang, D.N. | | Deposit date: | 2020-05-07 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
3CH8
 
 | | The crystal structure of PDZ-Fibronectin fusion protein | | Descriptor: | C-terminal octapeptide from protein ARVCF, MAGNESIUM ION, fusion protein PDZ-Fibronectin,Fibronectin | | Authors: | Makabe, K, Huang, J, Koide, A, Koide, S. | | Deposit date: | 2008-03-08 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for exquisite specificity of affinity clamps, synthetic binding proteins generated through directed domain-interface evolution.
J.Mol.Biol., 392, 2009
|
|
6D0N
 
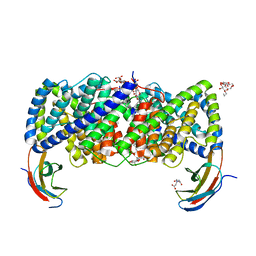 | | Crystal structure of a CLC-type fluoride/proton antiporter, V319G mutant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3UYO
 
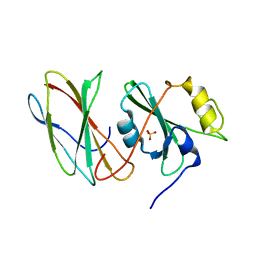 | |
7LO7
 
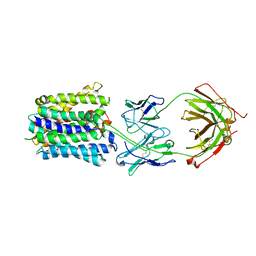 | | NorA in complex with Fab25 | | Descriptor: | Fab25 Heavy Chain, Fab25 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
7LO8
 
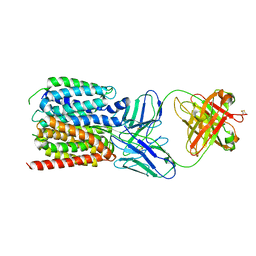 | | NorA in complex with Fab36 | | Descriptor: | Fab36 Heavy Chain, Fab36 Light Chain, Quinolone resistance protein NorA | | Authors: | Brawley, D.N, Sauer, D.B, Song, J.M, Koide, A, Koide, S, Traaseth, N.J, Wang, D.N. | | Deposit date: | 2021-02-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Nat.Chem.Biol., 18, 2022
|
|
6D0K
 
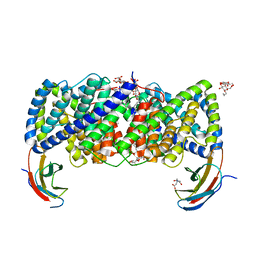 | | Crystal structure of a CLC-type fluoride/proton antiporter, E118Q mutant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6D0J
 
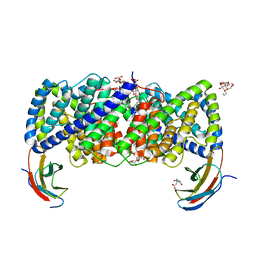 | | Crystal structure of a CLC-type fluoride/proton antiporter | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7JW7
 
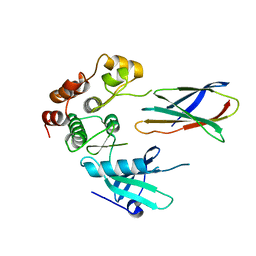 | | Structure of monobody 27 human MLKL pseudokinase domain complex | | Descriptor: | Mixed lineage kinase domain-like protein, Monobody 27 | | Authors: | Meng, Y, Garnish, S.E, Koide, A, Koide, S, Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2020-08-25 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Conformational interconversion of MLKL and disengagement from RIPK3 precede cell death by necroptosis.
Nat Commun, 12, 2021
|
|
8TTF
 
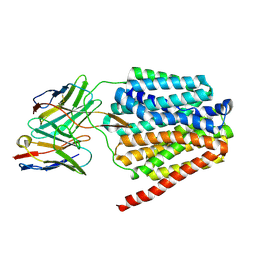 | | NorA double mutant - E222QD307N at pH 7.5 | | Descriptor: | Heavy Chain of FabDA1 Variable Domain, Light Chain of FabDA1 Variable Domain, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
7JXU
 
 | | Structure of monobody 32 human MLKL pseudokinase domain complex | | Descriptor: | 1,2-ETHANEDIOL, Mixed lineage kinase domain-like protein, Monobody 32 | | Authors: | Meng, Y, Garnish, S.E, Koide, A, Koide, S, Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Conformational interconversion of MLKL and disengagement from RIPK3 precede cell death by necroptosis.
Nat Commun, 12, 2021
|
|
8TTH
 
 | | NorA single mutant - D307N at pH 7.5 | | Descriptor: | Heavy Chain of FabDA1 Variable Domain, Light Chain of FabDA1 Variable Domain, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
8TTE
 
 | | Protonated state of NorA at pH 5.0 | | Descriptor: | FabDA1 CDRH3 loop, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
8TTG
 
 | | NorA single mutant - E222Q at pH 7.5 | | Descriptor: | FabDA1 CDRH3 loop, Quinolone resistance protein NorA | | Authors: | Li, J.P, Li, Y, Koide, A, Kuang, H.H, Torres, V.J, Koide, S, Wang, D.N, Traaseth, N.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Proton-coupled transport mechanism of the efflux pump NorA.
Nat Commun, 15, 2024
|
|
4IOF
 
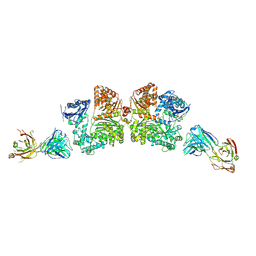 | | Crystal structure analysis of Fab-bound human Insulin Degrading Enzyme (IDE) | | Descriptor: | Fab-bound IDE, heavy chain, light chain, ... | | Authors: | McCord, L.A, Liang, W.G, Hoey, R, Dowdell, E, Koide, A, Koide, S, Tang, W.J. | | Deposit date: | 2013-01-07 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.353 Å) | | Cite: | Conformational states and recognition of amyloidogenic peptides of human insulin-degrading enzyme.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7KZX
 
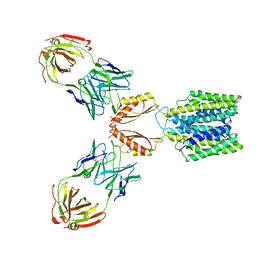 | | Cryo-EM structure of YiiP-Fab complex in Apo state | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab2R heavy chain, Fab2R light chain | | Authors: | Lopez-Redondo, M.L, Fan, S, Koide, A, Koide, S, Beckstein, O, Stokes, D.L. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Zinc binding alters the conformational dynamics and drives the transport cycle of the cation diffusion facilitator YiiP.
J.Gen.Physiol., 153, 2021
|
|
7KZZ
 
 | | Cryo-EM structure of YiiP-Fab complex in Holo state | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab2R heavy chain, Fab2R light chain, ... | | Authors: | Lopez-Redondo, M.L, Fan, S, Koide, A, Koide, S, Beckstein, O, Stokes, D.L. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Zinc binding alters the conformational dynamics and drives the transport cycle of the cation diffusion facilitator YiiP.
J.Gen.Physiol., 153, 2021
|
|
5V7P
 
 | | Atomic structure of the eukaryotic intramembrane Ras methyltransferase ICMT (isoprenylcysteine carboxyl methyltransferase), in complex with a monobody | | Descriptor: | DECANE, Protein-S-isoprenylcysteine O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Long, S.B, Diver, M.M, Pedi, L, Koide, A, Koide, S. | | Deposit date: | 2017-03-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic structure of the eukaryotic intramembrane RAS methyltransferase ICMT.
Nature, 553, 2018
|
|
