1VED
 
 | | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.9 angstroms resolution in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-30 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.9 angstroms resolution in space
To be Published
|
|
1VDS
 
 | | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.6 angstroms resolution in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the tetragonal form of hen egg white lysozyme at 1.6 angstroms resolution in space
to be published
|
|
1GYZ
 
 | |
6KMF
 
 | | FimA type V pilus from P.gingivalis | | Descriptor: | Major fimbrium subunit FimA type-1 | | Authors: | Shibata, S, Shoji, M, Matsunami, H, Matthews, M, Imada, K, Nakayama, K, Wolf, M. | | Deposit date: | 2019-07-31 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
1VDP
 
 | | The crystal structure of the monoclinic form of hen egg white lysozyme at 1.7 angstroms resolution in space | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, DeLucas, L.J, Hirose, M. | | Deposit date: | 2004-03-24 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the monoclinic form of hen egg white lysozyme at 1.7 angstroms resolution in space
to be published
|
|
2ZQ4
 
 | | The crystal structure of the orthorhombic form of hen egg white lysozyme at 2.0 angstroms resolution | | Descriptor: | Lysozyme C | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Yamane, T, Hirose, M. | | Deposit date: | 2008-08-03 | | Release date: | 2008-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.5 angstroms resolution
To be Published
|
|
2ZQ3
 
 | | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.6 angstroms resolution | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Aibara, S, Suzuki, A, Kidera, A, Shibata, K, Hirose, M. | | Deposit date: | 2008-08-03 | | Release date: | 2008-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the orthorhombic form of hen egg white lysozyme at 1.5 angstroms resolution
To be Published
|
|
1A7V
 
 | | CYTOCHROME C' FROM RHODOPSEUDOMONAS PALUSTRIS | | Descriptor: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shibata, N, Iba, S, Misaki, S, Meyer, T.E, Bartsch, R.G, Cusanovich, M.A, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1998-03-18 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Basis for monomer stabilization in Rhodopseudomonas palustris cytochrome c' derived from the crystal structure.
J.Mol.Biol., 284, 1998
|
|
7NVR
 
 | | Human Mediator with RNA Polymerase II Pre-initiation complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Rengachari, S, Schilbach, S, Aibara, S, Cramer, P. | | Deposit date: | 2021-03-15 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
7O4L
 
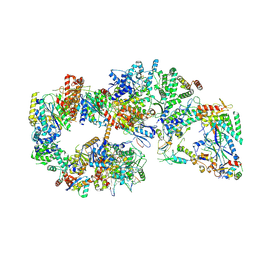 | | Yeast TFIIH in the expanded state within the pre-initiation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | Deposit date: | 2021-04-06 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
7O4J
 
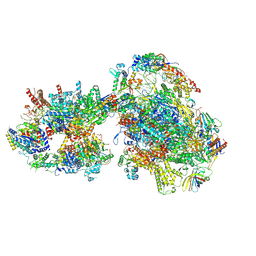 | | Yeast RNA polymerase II transcription pre-initiation complex (consensus) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | Deposit date: | 2021-04-06 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
7O4I
 
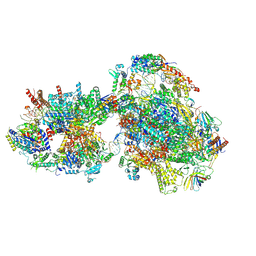 | | Yeast RNA polymerase II transcription pre-initiation complex with initial transcription bubble | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | Deposit date: | 2021-04-06 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
7O73
 
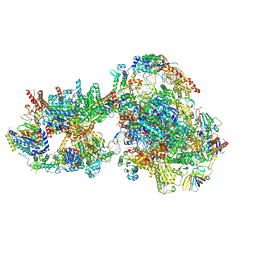 | | Yeast RNA polymerase II transcription pre-initiation complex with closed distorted promoter DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
7O75
 
 | | Yeast RNA polymerase II transcription pre-initiation complex with open promoter DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | Deposit date: | 2021-04-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
7O4K
 
 | | Yeast TFIIH in the contracted state within the pre-initiation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | Deposit date: | 2021-04-06 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
7O72
 
 | | Yeast RNA polymerase II transcription pre-initiation complex with closed promoter DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Schilbach, S, Aibara, S, Dienemann, C, Grabbe, F, Cramer, P. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of RNA polymerase II pre-initiation complex at 2.9 angstrom defines initial DNA opening.
Cell, 184, 2021
|
|
7B0Y
 
 | | Structure of a transcribing RNA polymerase II-U1 snRNP complex | | Descriptor: | 145-nt RNA, DNA-directed RNA polymerase II subunit D, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Zhang, S, Aibara, S, Vos, S.M, Agafonov, D.E, Luehrmann, R, Cramer, P. | | Deposit date: | 2020-11-23 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a transcribing RNA polymerase II-U1 snRNP complex.
Science, 371, 2021
|
|
7AOI
 
 | | Trypanosoma brucei mitochondrial ribosome large subunit assembly intermediate | | Descriptor: | 50S ribosomal protein L13, 50S ribosomal protein L14, 50S ribosomal protein L17, ... | | Authors: | Tobiasson, V, Gahura, O, Aibara, S, Baradaran, R, Zikova, A, Amunts, A. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-02 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Interconnected assembly factors regulate the biogenesis of mitoribosomal large subunit.
Embo J., 40, 2021
|
|
7B7U
 
 | | Cryo-EM structure of mammalian RNA polymerase II in complex with human RPAP2 | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Fianu, I, Dienemann, C, Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of mammalian RNA polymerase II in complex with human RPAP2.
Commun Biol, 4, 2021
|
|
4XWG
 
 | | Crystal Structure of LCAT (C31Y) in complex with Fab1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1 Heavy Chain, Fab1 Light Chain, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T. | | Deposit date: | 2015-01-28 | | Release date: | 2015-07-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The high-resolution crystal structure of human LCAT.
J.Lipid Res., 56, 2015
|
|
6I9R
 
 | | Large subunit of the human mitochondrial ribosome in complex with Virginiamycin M and Quinupristin | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Modelska, A, Aibara, S, Amunts, A. | | Deposit date: | 2018-11-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Inhibition of mitochondrial translation suppresses glioblastoma stem cell growth.
Cell Rep, 35, 2021
|
|
4XX1
 
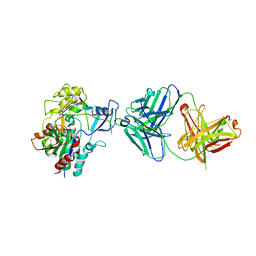 | | Low resolution structure of LCAT in complex with Fab1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1 heavy chain, Fab1 light chain, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T. | | Deposit date: | 2015-01-29 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The high-resolution crystal structure of human LCAT.
J.Lipid Res., 56, 2015
|
|
2KUP
 
 | | Solution structure of the complex of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of HALK | | Descriptor: | 19-residue peptide from ALK tyrosine kinase receptor, Fibroblast growth factor receptor substrate 3 | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
2KUQ
 
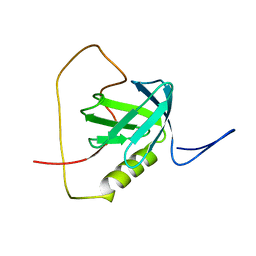 | | Solution structure of the chimera of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of HALK | | Descriptor: | Fibroblast growth factor receptor substrate 3,LINKER,ALK tyrosine kinase receptor | | Authors: | Li, H, Koshiba, S, Tomizawa, T, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
3J9W
 
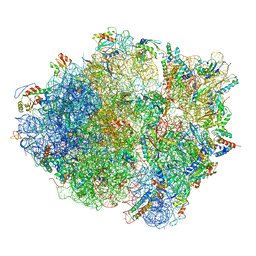 | | Cryo-EM structure of the Bacillus subtilis MifM-stalled ribosome complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein bS16, ... | | Authors: | Sohmen, D, Chiba, S, Shimokawa-Chiba, N, Innis, C.A, Berninghausen, O, Beckmann, R, Ito, K, Wilson, D.N. | | Deposit date: | 2015-03-16 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling.
Nat Commun, 6, 2015
|
|
